| Author(s) |
|
Posted on: 12 July 2024 purlPURL: https://gxy.io/GTN:N00094
We are proud to announce that a new training, explaining the analysis of single cell ATAC-seq data with SnapATAC2 and Scanpy, is now available in the Galaxy Training Network.
 Open image in new tab
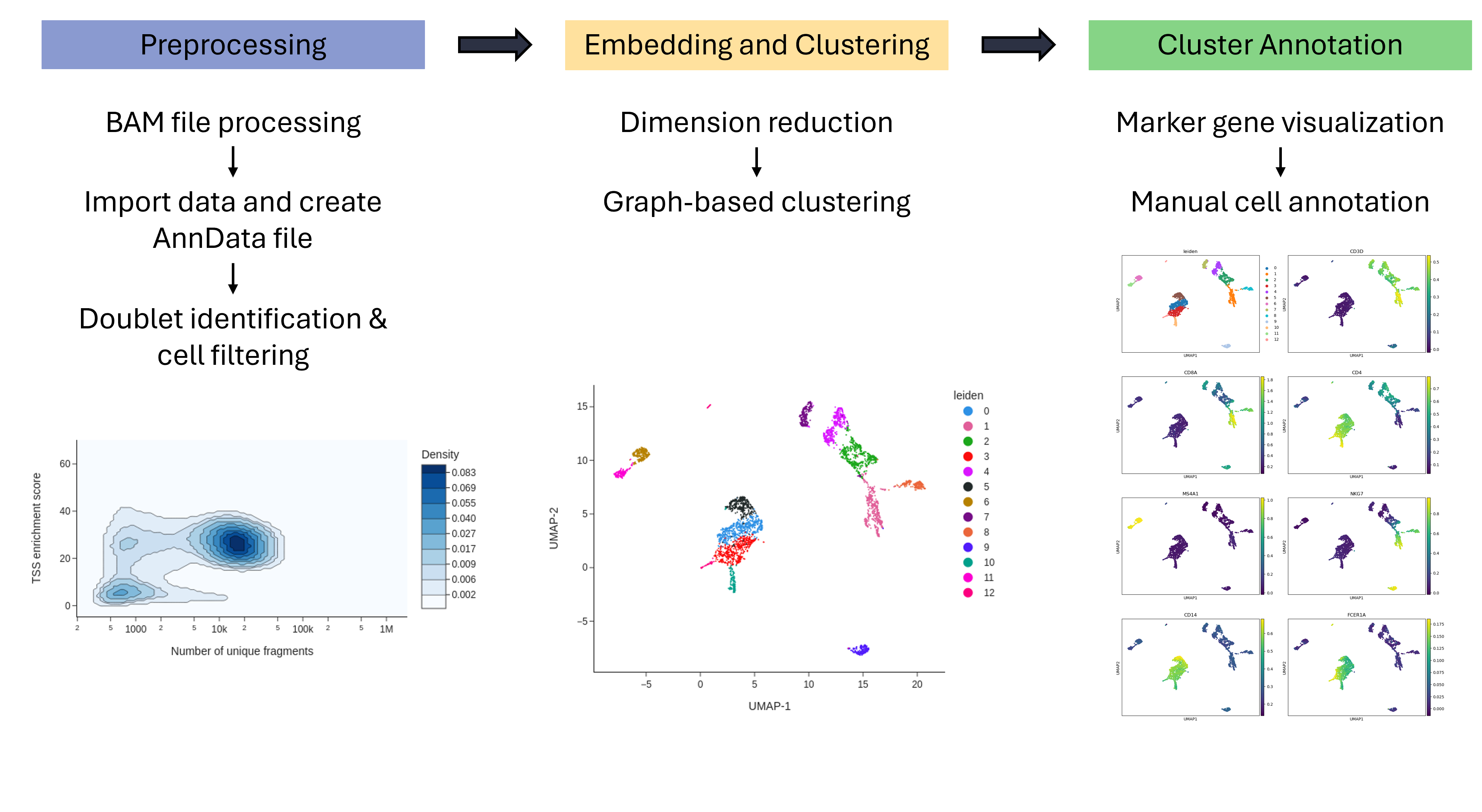
Open image in new tabThe tutorial consists of three sections: Preprocessing, Dimensionality Reduction and Clustering, and Cluster Annotation. During the preprocessing, cell-by-feature count matrices are created, which are filtered to produce high-quality AnnData count matrices. After that, the dimensionality of the data is reduced through nonlinear spectral embedding. The data is then projected onto two-dimensional space and clusters are identified. To annotate the clusters, the gene activity for each cell is measured and the activity of marker genes is visualized with the Scanpy package. The activity profile of each cluster is then used to determine the correct cell type.
View Material