name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/galaxy-interface" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/galaxy-interface/tutorials/upload-data-to-ena/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # Submitting SARS-CoV-2 sequences to ENA <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/roncoronimiguel/" class="contributor-badge contributor-roncoronimiguel"><img src="https://avatars.githubusercontent.com/roncoronimiguel?s=36" alt="Miguel Roncoroni avatar" width="36" class="avatar" /> Miguel Roncoroni</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00064">gxy.io/GTN:S00064</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) - Galaxy Basics for genomics: [<i class="fas fa-laptop" aria-hidden="true"></i><span class="visually-hidden">tutorial</span> hands-on](/training-material/topics/introduction/tutorials/galaxy-intro-101/tutorial.html) --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Introduce the European Nucleotide Archive (ENA) - Learn the requirements to submit raw SARS-CoV-2 sequences to ENA in Galaxy - Overview ENA's metadata model and how metadata objects are linked --- ### The European Nucleotide Archive .pull-left[ .left[ ENA is: - a FAIR and Open repository for sequence data (reads, assemblies, annotations) - part of the International Nucleotide Sequence Database Collaboration (INSDC) with NCBI and DDJB - the [COVID-19 data portal](https://www.covid19dataportal.org/) repository for SARS-CoV-2 sequences ] ] .pull-right[  The European Nucleotide Archive and INSDC ] --- ## SARS-CoV-2 sequences .left[ Why is raw SARS-CoV-2 sequence data important? - Allows reuse of data and reproducibility of analysis - Enables discovery of minor allelic variants and [intrahost variation](https://virological.org/t/global-framework-for-sars-cov-2-data-analysis-application-to-intrahost-variation-part-1/623) ] .image-40[  ] .reduce70[Minor allelic-variants can be used to detect intrahost variation. From Maier et al., 2021 [doi.org/10.1101/2021.03.25.437046](https://doi.org/10.1101/2021.03.25.437046)] --- ### Submitting reads with Galaxy .left[ Why use Galaxy to submit to ENA? - intuitive graphical user interface (GUI) - simple metadata input via a template spreadsheet or interactively - no bioinformatics skills needed ] .image-75[ ] --- ### Submission overview .image-100[  ] --- ### What you need .left[ Data: - compressed fastq format (*.fastq.gz, *.fastq.bz2) - human traces removed ([tutorial](https://training.galaxyproject.org/training-material/topics/sequence-analysis/tutorials/human-reads-removal/tutorial.html)) Metadata: - interactive metadata input (for a few submissions) or; - metadata [template spreadsheet](https://drive.google.com/file/d/1Gx78GKh58PmRjdmJ05DBbpObAL-3oUFX/view?usp=sharing) (for bulk submissions) Credentials: - [ENA Webin credentials](https://www.ebi.ac.uk/ena/submit/sra/#registration) in your Galaxy user information ] .left[  ] --- ### Metadata .left[ For the submission of SARS-CoV-2 reads [ENA's metadata model](https://ena-docs.readthedocs.io/en/latest/submit/general-guide/metadata.html) requires: - study, sample, experiment and run information - additional information for viral samples ([viral checklist](https://www.ebi.ac.uk/ena/browser/view/ERC000033)) ]  --- ### Metadata .left[ Interactive metadata input in Galaxy: ]  --- ### Metadata .left[ Metadata [template spreadsheet](https://drive.google.com/file/d/1Gx78GKh58PmRjdmJ05DBbpObAL-3oUFX/view?usp=sharing): - one sheet each for study, sample, experiment and run - built-in controlled vocabulary ]  --- ### Metadata .left[ - Different metadata objects are linked using Aliases - Aliases must be unique ]  --- ### Aliases .left[ Aliases link metadata objects: - Experiments are linked to Study and Samples - Runs are linked to Experiments ]  --- ### Aliases .left[ Aliases link metadata objects: - Experiments are linked to Study and Samples - Runs are linked to Experiments ]  --- ### Aliases .left[ Aliases link metadata to data: - Data (filename.fastq.gz) is linked to Run Alias ] 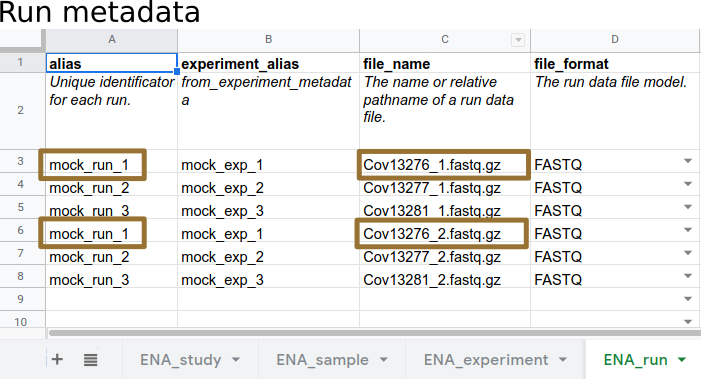 --- --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - ENA is a FAIR data repository for SARS-CoV-2 raw and assembled nucleotide data - You can easily submit reads to ENA using Galaxy's ENA upload tool (GUI, no bioinformatic skills needed) --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/roncoronimiguel/" class="contributor-badge contributor-roncoronimiguel"><img src="https://avatars.githubusercontent.com/roncoronimiguel?s=36" alt="Miguel Roncoroni avatar" width="36" class="avatar" /> Miguel Roncoroni</a> </td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-badge-small contributor-bgruening"><img src="https://avatars.githubusercontent.com/bgruening?s=36" alt="Björn Grüning avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/beatrizserrano/" class="contributor-badge contributor-badge-small contributor-beatrizserrano"><img src="https://avatars.githubusercontent.com/beatrizserrano?s=36" alt="Beatriz Serrano-Solano avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>