Identification of non-canonical ORFs and their potential biological function
Contributors
| Author(s) |
|
| Editor(s) |
|
Index of contents
-
Introduction
-
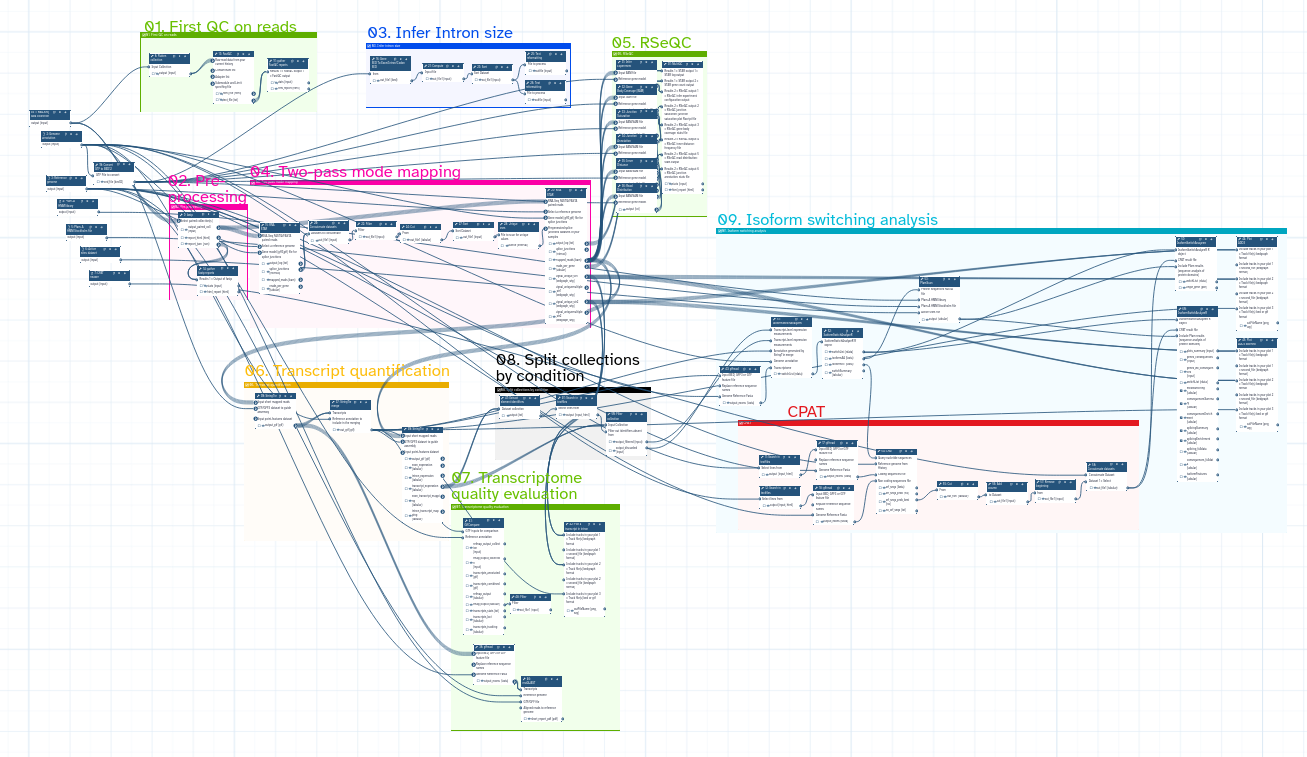
Galaxy workflow
Introduction
-
What do we mean by non-canonical ORFs?
-
Why study non-canonical ORFs?
-
Why have not been yet characterized?
-
How identify non-canonical ORFs?
What do we mean by non-canonical ORFs?

.footnote[Source: Cambridge dictionary]
What do we mean by non-canonical ORFs?
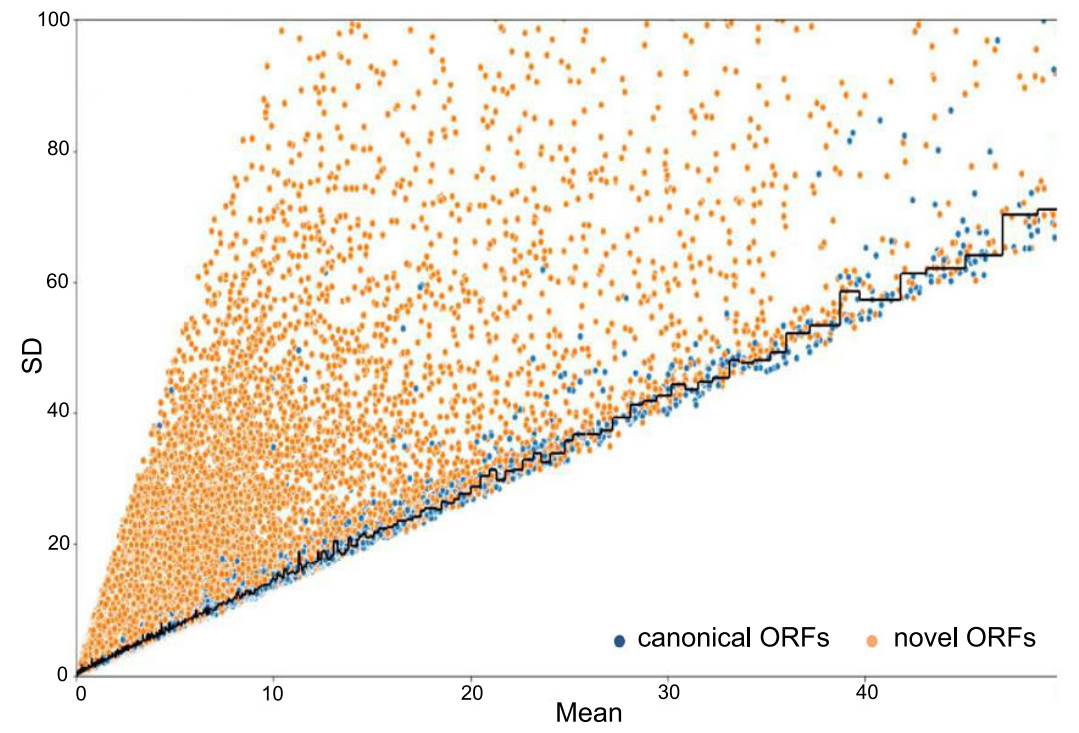
.footnote[Source: Erady et al. 2021]
What do we mean by non-canonical ORFs?

Why study non-canonical ORFs?
-
Potentially novel prognostic and diagnostic markers
-
The vast majority have not been investigated
-
Particulary attractive as allosteric celullar regulators
Why study non-canonical ORFs?
Why study non-canonical ORFs?
Why have not been characterized?
- Arbitrary thresholds on ORF lengths
- Peptides smaller than 100 aminoacids are usually discarted
- Frequently annotated as non-coding RNAs
- Propensity for structural disorder
- Discarted as intrinsically disordered proteins (IDPs)
Why study small peptides?

Why study small peptides?
Why study small peptides?
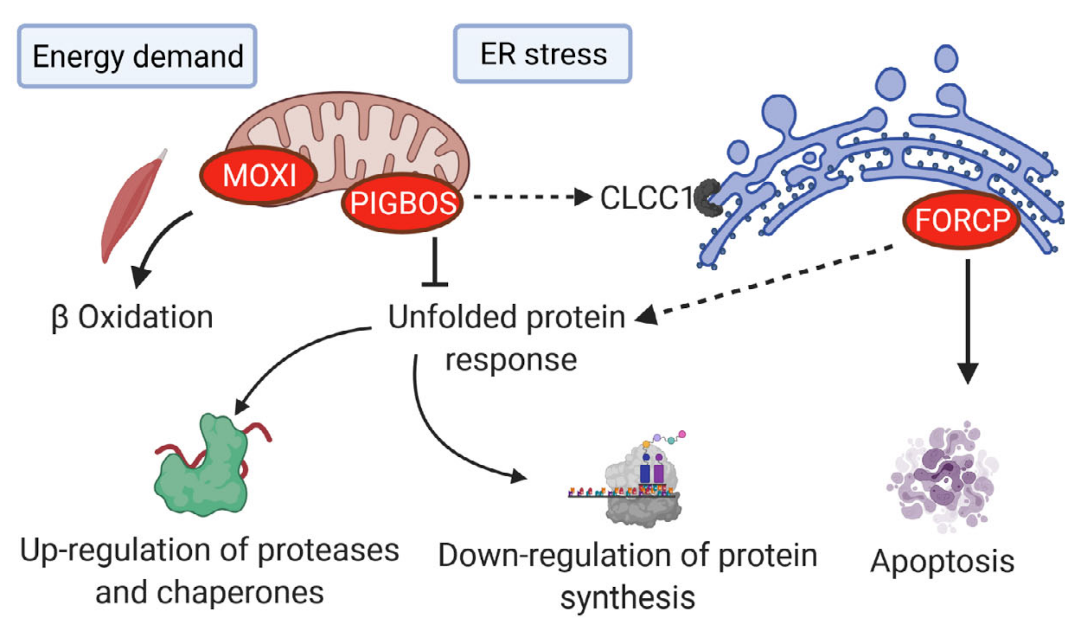
.footnote[Source: Steinberg and Koch 2021]
Annotated as non-coding RNAs?

Annotated as non-coding RNAs?

Annotated as non-coding RNAs?

Why study intrinsically disordered proteins (IDP)?
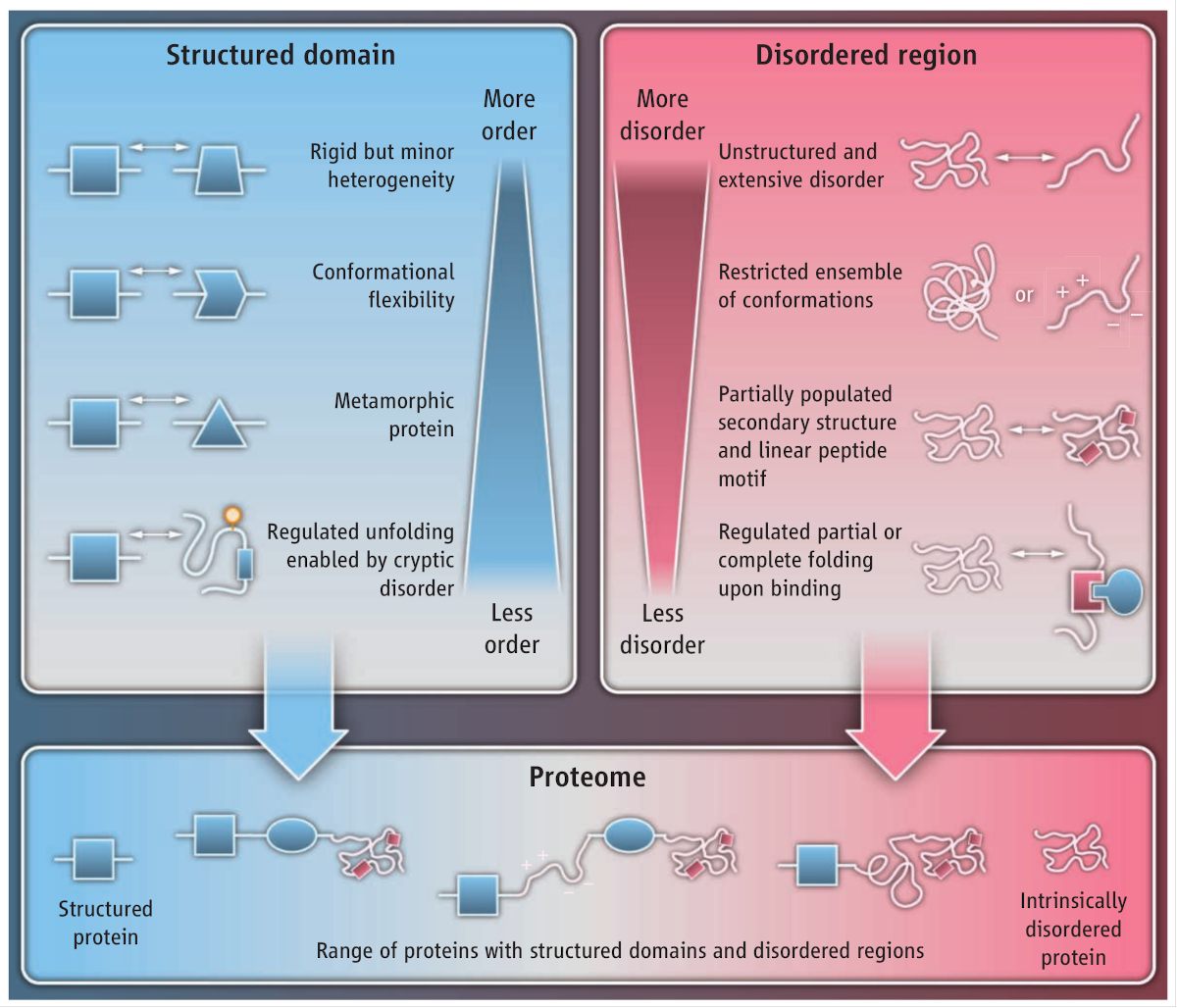
.footnote[Source: Babu et al. 2012]
Disorder-Function Paradigm
Disorder-Function Paradigm

Disorder-Function Paradigm
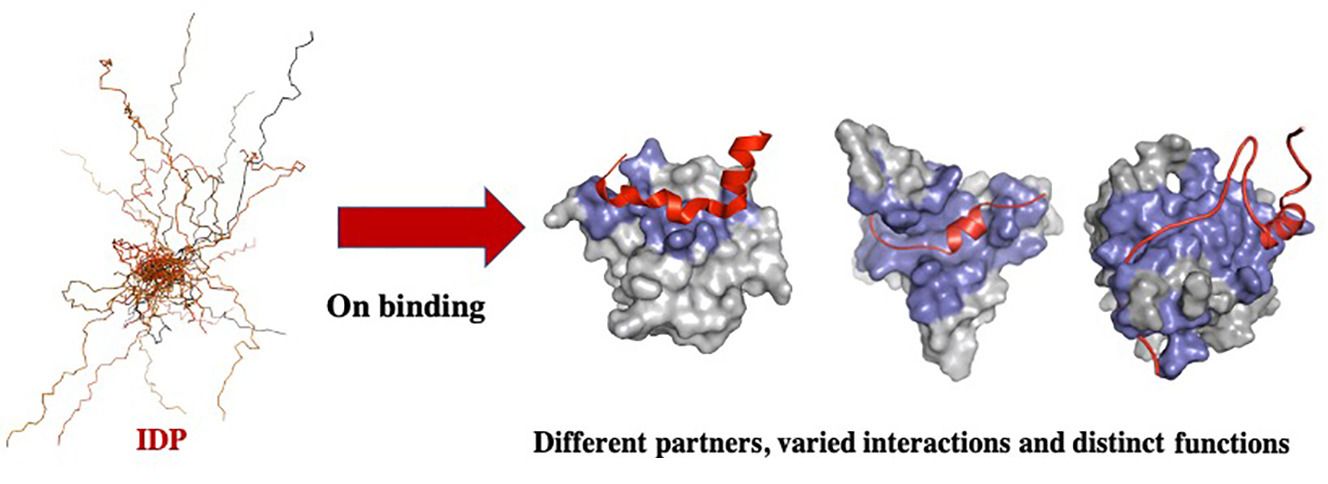
.footnote[Source: Chakrabarti and Chakravarty 2022]
How identify non-canonical ORFs?

How identify non-canonical ORFs?
How identify non-canonical ORFs?
Galaxy Workflow
Full detailed explanation in the Genome-wide alternative splicing analysis Galaxy training.
Galaxy Workflow
Full detailed explanation in the Genome-wide alternative splicing analysis Galaxy training.
Galaxy Workflow
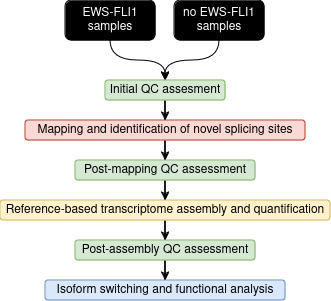
Initial QC assessment

Identify potential artifacts that may impact the interpretation of downstream analysis.
Mapping and identication of novel splicing sites with RNASTAR
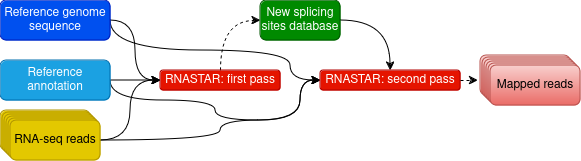
Two-pass alignment enables sequence reads to span novel splice junctions by fewer nucleotides, conferring greater read depth and providing significantly more accurate quantification of novel splice junctions.
Post-mapping QC assessment with RSeQC
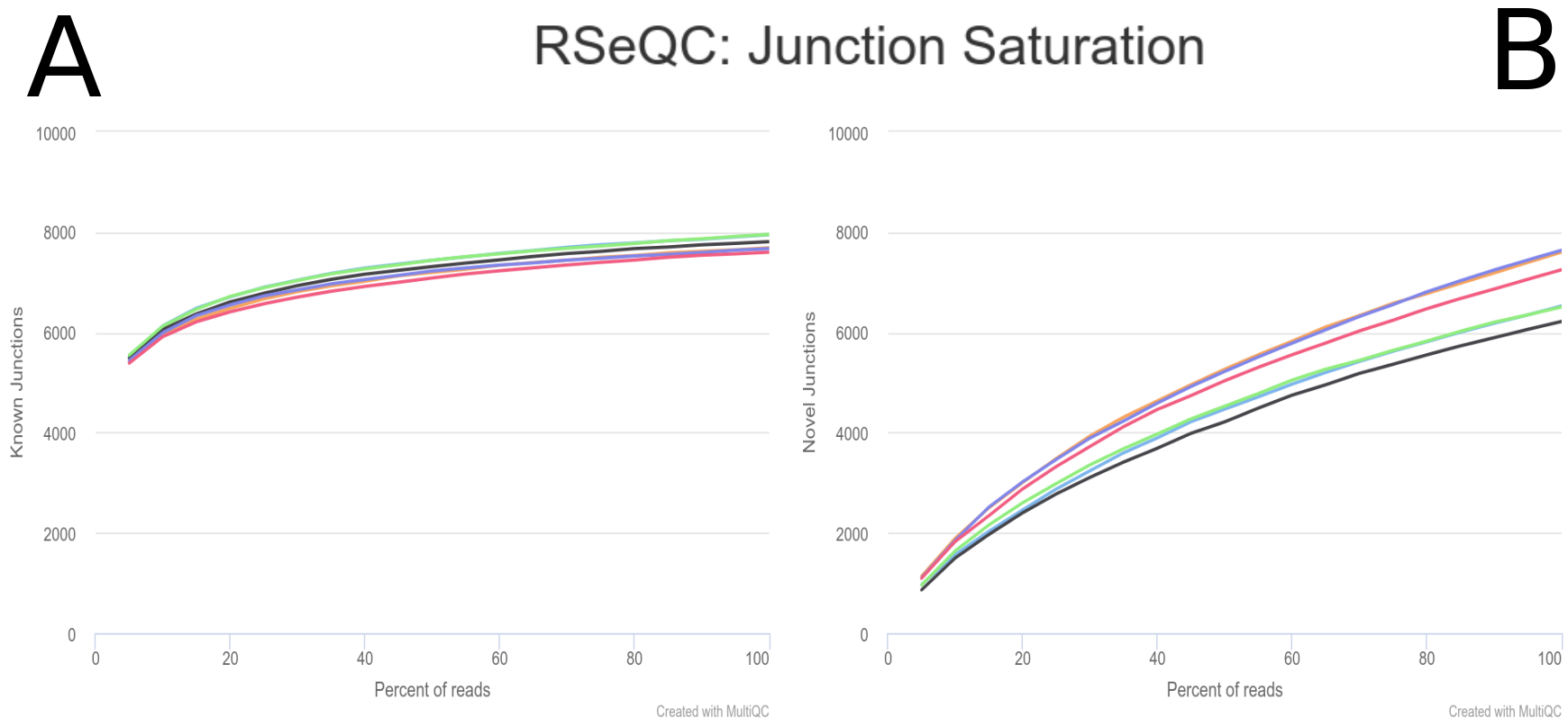
RSeQC is a toolkit for generating RNA-seq-specific quality control metrics. The figure corresponds to RSeQC junction saturation of known (A) and novel (B) splicing sites.
Reference-based transcriptome assembly and quantification with StringTie
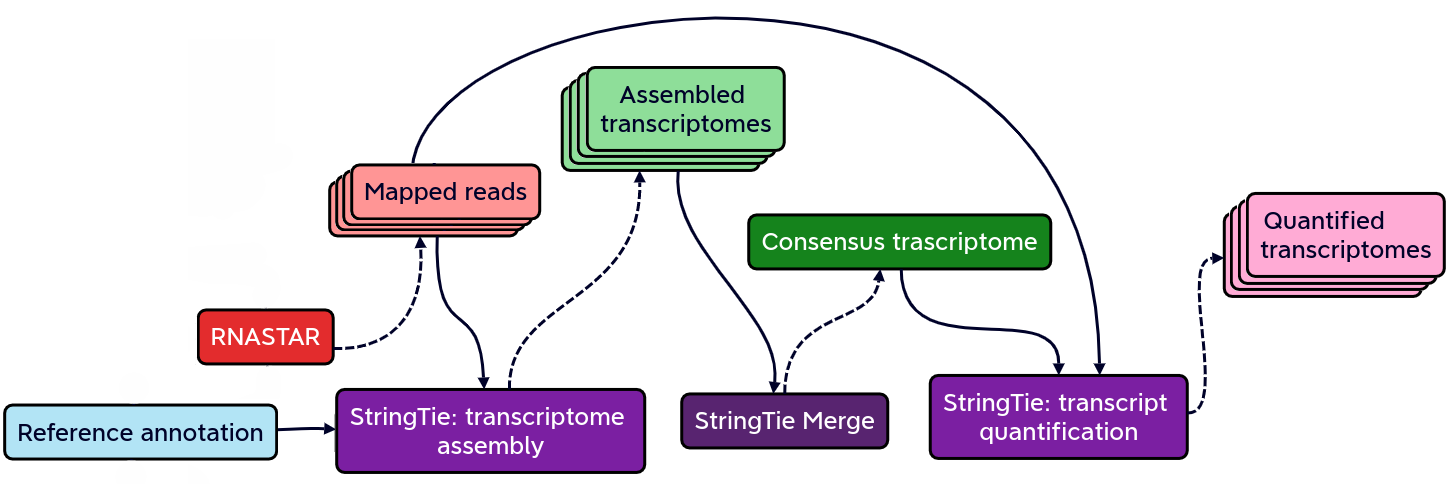
StringTie is a fast and highly efficient assembler of RNA-seq alignments into potential transcripts.
Post-assembly QC assessment with rnaQUAST
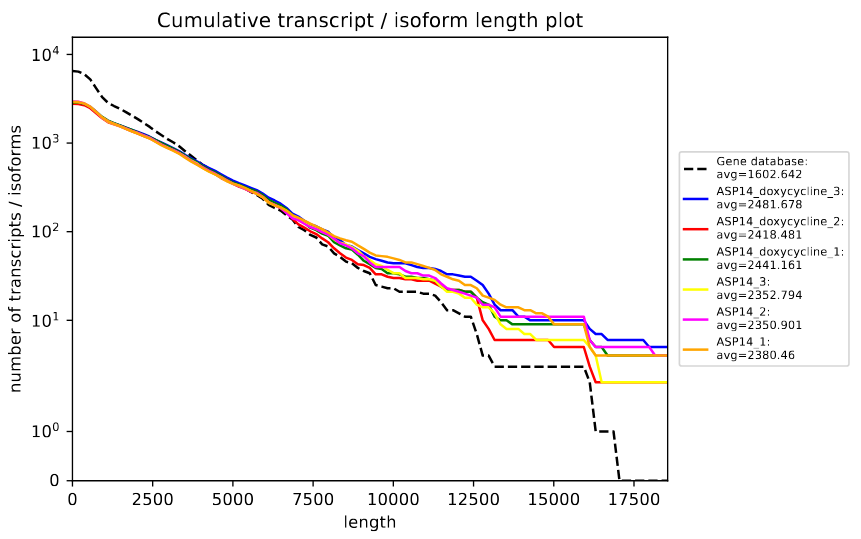
rnaQUAST, which will provide us diverse completeness/correctness statistics very useful in order to identify and address potential errors or gaps in the assembly process. The figure is a rnaQUAST cummulative isoform plot.
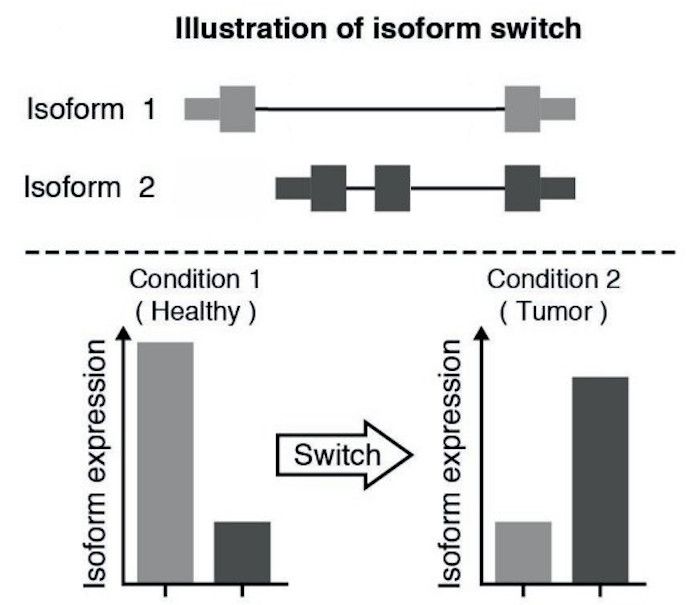
Isoform switching and functional analysis with IsoformSwitchAnalyzeR
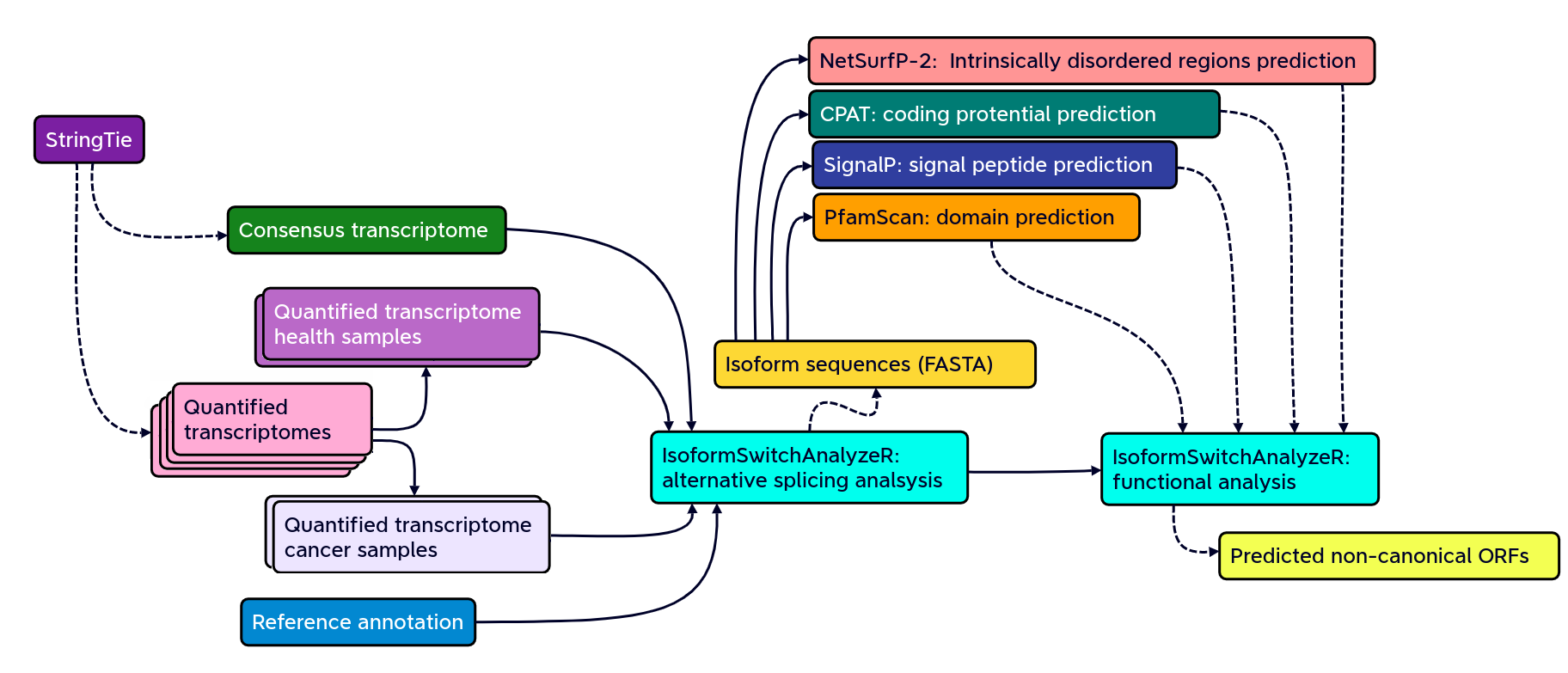
IsoformSwitchAnalyzieR performs the differential isoform usage analysis by using DEXSeq.
Isoform switching and functional analysis with IsoformSwitchAnalyzeR
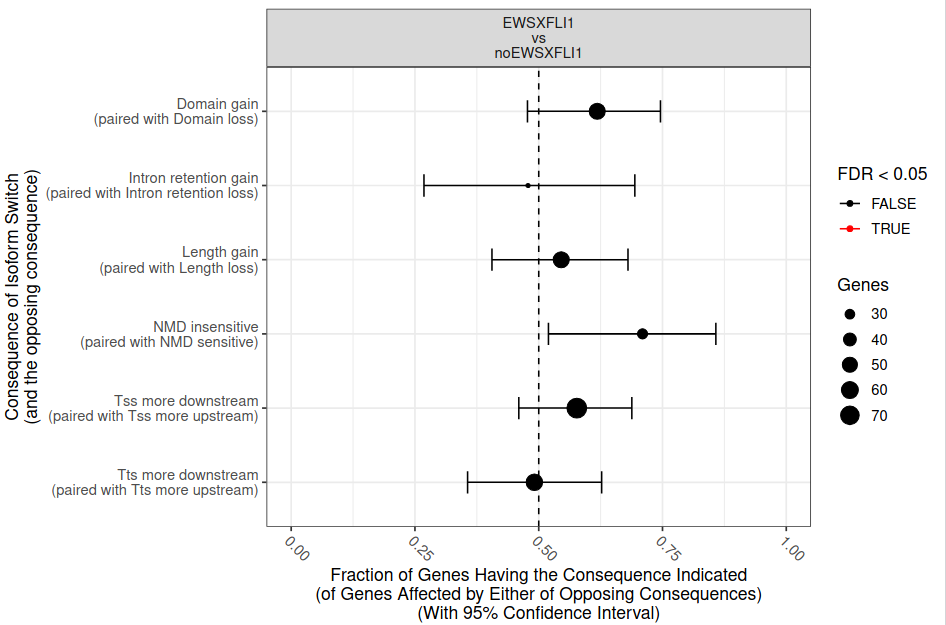
To analyze large-scale patterns in predicted IS consequences, IsoformSwitchAnalyzeR computes all isoform switching events resulting in a gain/loss of a specific consequence (e.g. protein domain gain/loss)
Thank you!
This material is the result of a collaborative work. Thanks to the Galaxy Training Network and all the contributors! Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.
Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.
References
- Babu, M. M., R. W. Kriwacki, and R. V. Pappu, 2012 Versatility from Protein Disorder. Science 337: 1460–1461. 10.1126/science.1228775
- Erady, C., A. Boxall, S. Puntambekar, N. S. Jagannathan, R. Chauhan et al., 2021 Pan-cancer analysis of transcripts encoding novel open-reading frames (nORFs) and their potential biological functions. npj Genomic Medicine 6: 10.1038/s41525-020-00167-4
- Steinberg, R., and H.-G. Koch, 2021 The largely unexplored biology of small proteins in pro- and eukaryotes. The FEBS Journal 288: 7002–7024. 10.1111/febs.15845
- Chakrabarti, P., and D. Chakravarty, 2022 Intrinsically disordered proteins/regions and insight into their biomolecular interactions. Biophysical Chemistry 283: 106769. 10.1016/j.bpc.2022.106769