name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/transcriptomics" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/transcriptomics/tutorials/differential-isoform-expression/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # Identification of non-canonical ORFs and their potential biological function <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-gallardoalba"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /> Cristóbal Gallardo</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00108">gxy.io/GTN:S00108</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) - [Sequence analysis](/training-material/topics/sequence-analysis) - Quality Control: [<i class="fab fa-slideshare" aria-hidden="true"></i><span class="visually-hidden">slides</span> slides](/training-material/topics/sequence-analysis/tutorials/quality-control/slides.html) - [<i class="fas fa-laptop" aria-hidden="true"></i><span class="visually-hidden">tutorial</span> hands-on](/training-material/topics/sequence-analysis/tutorials/quality-control/tutorial.html) - Mapping: [<i class="fab fa-slideshare" aria-hidden="true"></i><span class="visually-hidden">slides</span> slides](/training-material/topics/sequence-analysis/tutorials/mapping/slides.html) - [<i class="fas fa-laptop" aria-hidden="true"></i><span class="visually-hidden">tutorial</span> hands-on](/training-material/topics/sequence-analysis/tutorials/mapping/tutorial.html) --- ## Index of contents 1. Introduction 2. Galaxy workflow --- ## Introduction - What do we mean by non-canonical ORFs? - Why study non-canonical ORFs? - Why have not been yet characterized? - How identify non-canonical ORFs? --- ## What do we mean by non-canonical ORFs?  .footnote[Source: [Cambridge dictionary](https://dictionary.cambridge.org/dictionary/english/canonical)] --- ## What do we mean by non-canonical ORFs?  .footnote[Source: <span class="citation"><a href="https://doi.org/10.1038/s41525-020-00167-4">Erady <i>et al.</i> 2021</a></span>] --- ## What do we mean by non-canonical ORFs?  --- ## Why study non-canonical ORFs? - Potentially novel <b>prognostic and diagnostic markers</b> - The vast majority <b>have not been investigated</b> - Particulary attractive as <b>allosteric celullar regulators</b> --- ## Why study non-canonical ORFs?  --- ## Why study non-canonical ORFs?  --- ## Why have not been characterized? - Arbitrary thresholds on ORF lengths - Peptides smaller than 100 aminoacids are usually discarted - Frequently annotated as non-coding RNAs - Propensity for structural disorder - Discarted as intrinsically disordered proteins (IDPs) --- ## Why study small peptides?  --- ## Why study small peptides?  --- ## Why study small peptides?  .footnote[Source: <span class="citation"><a href="https://doi.org/10.1111/febs.15845">Steinberg and Koch 2021</a></span>] --- ## Annotated as non-coding RNAs?  --- ## Annotated as non-coding RNAs?  --- ## Annotated as non-coding RNAs?  --- ## Why study intrinsically disordered proteins (IDP)?  .footnote[Source: <span class="citation"><a href="https://doi.org/10.1126/science.1228775">Babu <i>et al.</i> 2012</a></span>] --- ## Disorder-Function Paradigm  --- ## Disorder-Function Paradigm  --- ## Disorder-Function Paradigm  .footnote[Source: <span class="citation"><a href="https://doi.org/10.1016/j.bpc.2022.106769">Chakrabarti and Chakravarty 2022</a></span>] --- ## How identify non-canonical ORFs?  --- ## How identify non-canonical ORFs?  --- ## How identify non-canonical ORFs?  --- ## Galaxy Workflow  Full detailed explanation in the [Genome-wide alternative splicing analysis](/training-material/topics/transcriptomics/tutorials/differential-isoform-expression/tutorial.html) Galaxy training. --- ## Galaxy Workflow  Full detailed explanation in the [Genome-wide alternative splicing analysis](/training-material/topics/transcriptomics/tutorials/differential-isoform-expression/tutorial.html) Galaxy training. --- ## Galaxy Workflow 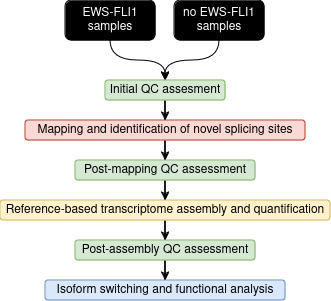 --- ## Initial QC assessment  Identify potential artifacts that may impact the interpretation of downstream analysis. --- ## Mapping and identication of novel splicing sites with RNASTAR 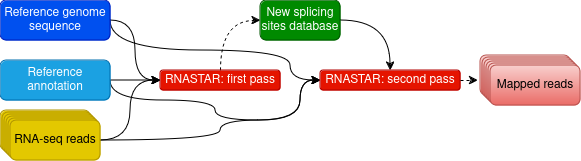 Two-pass alignment enables sequence reads to span novel splice junctions by fewer nucleotides, conferring greater read depth and providing significantly more accurate quantification of novel splice junctions. --- ## Post-mapping QC assessment with RSeQC 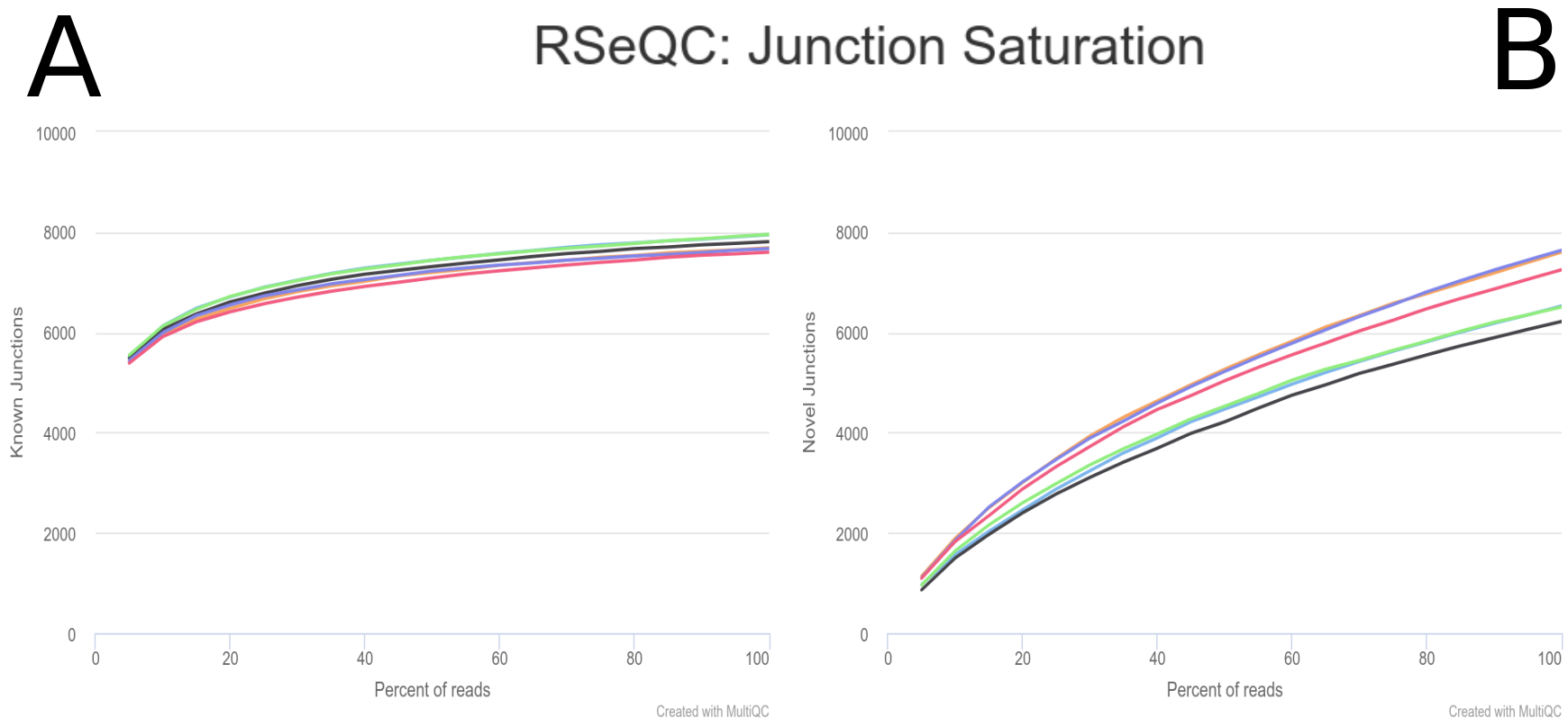 RSeQC is a toolkit for generating RNA-seq-specific quality control metrics. The figure corresponds to RSeQC junction saturation of known (A) and novel (B) splicing sites. --- ## Reference-based transcriptome assembly and quantification with StringTie 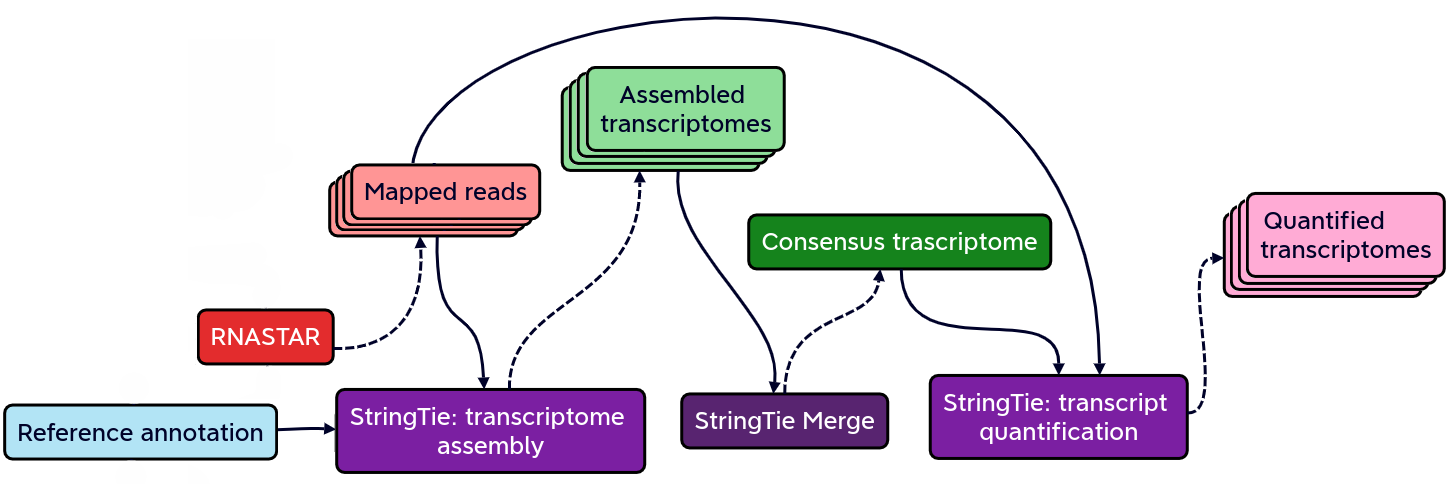 StringTie is a fast and highly efficient assembler of RNA-seq alignments into potential transcripts. --- ## Post-assembly QC assessment with rnaQUAST  rnaQUAST, which will provide us diverse completeness/correctness statistics very useful in order to identify and address potential errors or gaps in the assembly process. The figure is a rnaQUAST cummulative isoform plot. --- ## Isoform switching and functional analysis with IsoformSwitchAnalyzeR  IsoformSwitchAnalyzieR performs the differential isoform usage analysis by using DEXSeq. --- ## Isoform switching and functional analysis with IsoformSwitchAnalyzeR  To analyze large-scale patterns in predicted IS consequences, IsoformSwitchAnalyzeR computes all isoform switching events resulting in a gain/loss of a specific consequence (e.g. protein domain gain/loss) --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-gallardoalba"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /> Cristóbal Gallardo</a> </td> </tr> <tr> <td><abbr title="These people edited the text, either for spelling and grammar, flow, GTN-fit, or other similar editing categories">Editor(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/lldelisle/" class="contributor-badge contributor-lldelisle"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/lldelisle?s=36" alt="Lucille Delisle avatar" width="36" class="avatar" /> Lucille Delisle</a></td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-badge-small contributor-bgruening"><img src="https://avatars.githubusercontent.com/bgruening?s=36" alt="Björn Grüning avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/lldelisle/" class="contributor-badge contributor-badge-small contributor-lldelisle"><img src="https://avatars.githubusercontent.com/lldelisle?s=36" alt="Lucille Delisle avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-badge-small contributor-gallardoalba"><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-badge-small contributor-hexylena"><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/wm75/" class="contributor-badge contributor-badge-small contributor-wm75"><img src="https://avatars.githubusercontent.com/wm75?s=36" alt="Wolfgang Maier avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>