Helena Rasche
Affiliations
Former Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Galaxy Server administration
- Foundations of Data Science
- Development in Galaxy
- Using Galaxy and Managing your Data
- Genome Annotation
- GMOD
- One Health
- Visualisation
- Admin Training Course
- Gallantries Grant - Intellectual Output 1 - Introduction to data analysis and -management, statistics, and coding
- Gallantries Grant - Intellectual Output 2 - Large-scale data analysis, and introduction to visualisation and data modelling
- Gallantries Grant - Intellectual Output 3 - Data stewardship, federation, standardisation, and collaboration
- Introductory Python
- Introductory SQL
Tutorials
- Microbiome / Metatranscriptomics analysis using microbiome RNA-seq data 🧐
- Microbiome / Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition 🧐
- Microbiome / 16S Microbial Analysis with mothur (extended) 🧐
- Microbiome / Metatranscriptomics analysis using microbiome RNA-seq data (short) 🧐
- Microbiome / Identifying Mycorrhizal Fungi from ITS2 sequencing using LotuS2 🧐
- Microbiome / Taxonomic Profiling and Visualization of Metagenomic Data 🧐
- Microbiome / Analyses of metagenomics data - The global picture 🧐
- Microbiome / Assembly of metagenomic sequencing data 🧐
- Microbiome / Antibiotic resistance detection 🧐
- Microbiome / 16S Microbial Analysis with mothur (short) 🧐
- Microbiome / Building an amplicon sequence variant (ASV) table from 16S data using DADA2 🧐
- Microbiome / Identification of the micro-organisms in a beer using Nanopore sequencing 🧐
- Microbiome / Binning of metagenomic sequencing data 🧐
- Using Galaxy and Managing your Data / Automating Galaxy workflows using the command line 🧐
- Using Galaxy and Managing your Data / Understanding Galaxy history system 📝 🧐
- Using Galaxy and Managing your Data / Downloading and Deleting Data in Galaxy ✍️ 🧐
- Using Galaxy and Managing your Data / JupyterLab in Galaxy 📝 🧐
- Using Galaxy and Managing your Data / Extracting Workflows from Histories 🧐
- Using Galaxy and Managing your Data / Workflow Reports ⚙️ 🧐
- Using Galaxy and Managing your Data / Submitting sequence data to ENA 🧐
- Using Galaxy and Managing your Data / Creating, Editing and Importing Galaxy Workflows 🧐
- Using Galaxy and Managing your Data / InterMine integration with Galaxy 🧐
- Using Galaxy and Managing your Data / Rule Based Uploader ✍️ 🧐
- Using Galaxy and Managing your Data / SRA Aligned Read Format to Speed Up SARS-CoV-2 data Analysis 🧐
- Using Galaxy and Managing your Data / Use Jupyter notebooks in Galaxy 🧐
- Using Galaxy and Managing your Data / Name tags for following complex histories ✍️ 🧐
- Using Galaxy and Managing your Data / RStudio in Galaxy 🧐
- Using Galaxy and Managing your Data / Using dataset collections 🧐
- Using Galaxy and Managing your Data / Group tags for complex experimental designs 🧐
- Using Galaxy and Managing your Data / Rule Based Uploader: Advanced ✍️ 🧐
- Using Galaxy and Managing your Data / Creating high resolution images of Galaxy Workflows 📝 🧐
- Using Galaxy and Managing your Data / Using Workflow Parameters ✍️ 🧐
- Using Galaxy and Managing your Data / Searching Your History ✍️ 🧐
- Galaxy Server administration / Alternative Celery Deployment for Galaxy ✍️
- Galaxy Server administration / Customizing the look of Galaxy 📝 🧐
- Galaxy Server administration / Deploying a compute cluster in OpenStack via Terraform ✍️ 🧐
- Galaxy Server administration / Deploying Wireguard for private mesh networking ✍️ 🧐
- Galaxy Server administration / Enable upload via FTP 🧐
- Galaxy Server administration / Deploying a Beacon v1 in Galaxy ✍️ 🧐
- Galaxy Server administration / Create a subdomain for your community on UseGalaxy.eu 🧐
- Galaxy Server administration / Galaxy Database schema 🧐
- Galaxy Server administration / Galaxy Installation with Ansible ✍️ 🧐
- Galaxy Server administration / Server Maintenance: Cleanup, Backup, and Restoration ✍️ 🧐
- Galaxy Server administration / Pulsar usage on SURF Research Cloud ✍️ 🧐
- Galaxy Server administration / Galaxy Interactive Tools ✍️ 🧐
- Galaxy Server administration / Reference Data with CVMFS ✍️ 🧐
- Galaxy Server administration / Use Apptainer containers for running Galaxy jobs ✍️ 🧐
- Galaxy Server administration / Data Libraries ✍️ 🧐
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar ✍️ 🧐
- Galaxy Server administration / Ansible ✍️ 🧐
- Galaxy Server administration / Reference Data with Data Managers 📝 🧐
- Galaxy Server administration / Performant Uploads with TUS ✍️ 🧐
- Galaxy Server administration / Galaxy Monitoring with gxadmin ✍️ 🧐
- Galaxy Server administration / External Authentication ✍️ 🧐
- Galaxy Server administration / Customizing the look of Galaxy (Manual) 📝 🧐
- Galaxy Server administration / Distributed Object Storage ✍️ 🧐
- Galaxy Server administration / Managing Galaxy on Kubernetes 🧐
- Galaxy Server administration / Mapping Jobs to Destinations using TPV 📝 🧐
- Galaxy Server administration / Setting up Celery Workers for Galaxy 📝 🧐
- Galaxy Server administration / Monitoring Galaxy and Pulsar with Sentry 📝 🧐
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana ✍️ 🧐
- Galaxy Server administration / Reference Data with CVMFS without Ansible ✍️ 🧐
- Galaxy Server administration / Training Infrastructure as a Service (TIaaS) ✍️ 🧐
- Galaxy Server administration / Automation with Jenkins ✍️ 🧐
- Galaxy Server administration / Galaxy Tool Management with Ephemeris ✍️ 🧐
- Galaxy Server administration / Connecting Galaxy to a compute cluster ✍️ 🧐
- Galaxy Server administration / Galaxy usage on SURF Research Cloud ✍️ 🧐
- Galaxy Server administration / Galaxy Installation on Kubernetes 🧐
- Galaxy Server administration / Deploying Tailscale/Headscale for private mesh networking ✍️ 🧐
- Galaxy Server administration / Upgrading Galaxy 🧐
- Galaxy Server administration / How I learned to stop worrying and love the systemd ✍️
- Galaxy Server administration / Galaxy Monitoring with Reports ✍️ 🧐
- Visualisation / Genomic Data Visualisation with JBrowse ✍️ 🧐
- Visualisation / Ploting a Microbial Genome with Circos ✍️
- Visualisation / Visualisation with Circos ✍️ 🧐
- Single Cell / Importing files from public atlases 🧐
- Single Cell / Filter, plot, and explore single cell RNA-seq data with Seurat 📝 🧐
- Single Cell / Filter, plot, and explore single cell RNA-seq data with Seurat (R) 📝 🧐
- Single Cell / Filter, plot and explore single-cell RNA-seq data with Scanpy 📝 🧐
- Single Cell / Combining single cell datasets after pre-processing 📝 🧐
- Single Cell / Removing the effects of the cell cycle 🧐
- Single Cell / Analysis of plant scRNA-Seq Data with Scanpy 🧐
- Single Cell / Pre-processing of 10X Single-Cell ATAC-seq Datasets 🧐
- Single Cell / Scanpy Parameter Iterator 🧐
- Single Cell / Inferring single cell trajectories with Monocle3 📝 🧐
- Single Cell / Downstream Single-cell RNA analysis with RaceID 🧐
- Single Cell / Inferring single cell trajectories with Scanpy (Python) 📝 🧐
- Single Cell / Converting NCBI Data to the AnnData Format 📝 🧐
- Single Cell / Pre-processing of Single-Cell RNA Data 🧐
- Single Cell / Generating a single cell matrix using Alevin 📝 🧐
- Single Cell / Single-cell quality control with scater 🧐
- Single Cell / Converting between common single cell data formats 📝 🧐
- Single Cell / Bulk RNA Deconvolution with MuSiC 🧐
- Single Cell / Generating a single cell matrix using Alevin and combining datasets (bash + R) 🧐
- Single Cell / Comparing inferred cell compositions using MuSiC deconvolution 🧐
- Single Cell / Inferring single cell trajectories with Scanpy 📝 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy 🧐
- Single Cell / Matrix Exchange Format to ESet | Creating a single-cell RNA-seq reference dataset for deconvolution 🧐
- Single Cell / Pre-processing of 10X Single-Cell RNA Datasets 🧐
- Single Cell / Bulk matrix to ESet | Creating the bulk RNA-seq dataset for deconvolution 🧐
- Single Cell / Inferring single cell trajectories with Monocle3 (R) 📝 🧐
- Single Cell / Filter, plot and explore single-cell RNA-seq data with Scanpy (Python) 🧐
- Single Cell / Understanding Barcodes 🧐
- Single Cell / GO Enrichment Analysis on Single-Cell RNA-Seq Data 🧐
- Computational chemistry / Protein-ligand docking 🧐
- Computational chemistry / Virtual screening of the SARS-CoV-2 main protease with rxDock and pose scoring 🧐
- Computational chemistry / Data management in Medicinal Chemistry 🧐
- Computational chemistry / Protein target prediction of a bioactive ligand with Align-it and ePharmaLib 🧐
- Computational chemistry / Running molecular dynamics simulations using NAMD 🧐
- Computational chemistry / Analysis of molecular dynamics simulations 🧐
- Computational chemistry / Setting up molecular systems 🧐
- Computational chemistry / High Throughput Molecular Dynamics and Analysis 🧐
- Computational chemistry / Running molecular dynamics simulations using GROMACS 🧐
- Teaching and Hosting Galaxy training / Teaching experiences 🧐
- Teaching and Hosting Galaxy training / Assessment and feedback in training and teachings 🧐
- Teaching and Hosting Galaxy training / Course Builder ✍️ 🧐
- Teaching and Hosting Galaxy training / Training Infrastructure as a Service ✍️ 🧐
- Teaching and Hosting Galaxy training / Set up a Galaxy for Training 🧐
- Teaching and Hosting Galaxy training / Training techniques to enhance learner participation and engagement 🧐
- Teaching and Hosting Galaxy training / Motivation and Demotivation 🧐
- Teaching and Hosting Galaxy training / Organizing a workshop ✍️
- Teaching and Hosting Galaxy training / Galaxy Admin Training ✍️ 🧐
- Teaching and Hosting Galaxy training / Running a workshop as instructor ✍️ 🧐
- Teaching and Hosting Galaxy training / Asynchronous training ✍️ 🧐
- Synthetic Biology / Evaluating and ranking a set of pathways based on multiple metrics 🧐
- Synthetic Biology / Designing plasmids encoding predicted pathways by using the BASIC assembly method 🧐
- Ecology / QGIS Web Feature Services 🧐
- Ecology / Species distribution modeling 🧐
- Ecology / RAD-Seq Reference-based data analysis 🧐
- Ecology / Compute and analyze biodiversity metrics with PAMPA toolsuite 🧐
- Ecology / Metabarcoding/eDNA through Obitools 🧐
- Ecology / Checking expected species and contamination in bacterial isolate 🧐
- Ecology / Obis marine indicators 🧐
- Ecology / RAD-Seq to construct genetic maps 🧐
- Ecology / Biodiversity data exploration 🧐
- Ecology / From NDVI data with OpenEO to time series visualisation with Holoviews 🧐
- Ecology / Sentinel 2 biodiversity 🧐
- Ecology / RAD-Seq de-novo data analysis 🧐
- Ecology / Visualization of Climate Data using NetCDF xarray Map Plotting 🧐
- Ecology / Regional GAM 🧐
- Ecology / Creating metadata using Ecological Metadata Language (EML) standard with EML Assembly Line functionalities 📝 🧐
- Ecology / Preparing genomic data for phylogeny reconstruction 🧐
- Ecology / Visualize EBV cube data with Panoply netCDF viewer 🧐
- Ecology / Ecoregionalization workflow tutorial 🧐
- Ecology / Creating FAIR Quality assessment reports and draft of Data Papers from EML metadata with MetaShRIMPS 📝 🧐
- Ecology / Cleaning GBIF data for the use in Ecology 🧐
- Ecology / Champs blocs indicators 🧐
- Assembly / Genome assembly using PacBio data 🧐
- Assembly / De Bruijn Graph Assembly ✍️ 🧐
- Assembly / Making sense of a newly assembled genome ✍️ 🧐
- Assembly / Using the VGP workflows to assemble a vertebrate genome with HiFi and Hi-C data 🧐
- Assembly / Chloroplast genome assembly 🧐
- Assembly / Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data 📝 🧐
- Assembly / Genome Assembly Quality Control 🧐
- Assembly / Large genome assembly and polishing 🧐
- Assembly / Genome Assembly of MRSA from Oxford Nanopore MinION data (and optionally Illumina data) 📝 🧐
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads 🧐
- Assembly / Vertebrate genome assembly using HiFi, Bionano and Hi-C data - Step by Step 🧐
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / Unicycler Assembly 🧐
- Climate / Getting your hands-on climate data 🧐
- Climate / Sentinel 5P data visualisation 🧐
- Climate / Ocean's variables study 🧐
- Climate / Visualize Climate data with Panoply netCDF viewer 🧐
- Climate / Pangeo ecosystem 101 for everyone - Introduction to Xarray Galaxy Tools 🧐
- Climate / Ocean Data View (ODV) 🧐
- Climate / Functionally Assembled Terrestrial Ecosystem Simulator (FATES) with Galaxy Climate JupyterLab 🧐
- Climate / Functionally Assembled Terrestrial Ecosystem Simulator (FATES) 🧐
- Climate / Getting your hands-on earth data 🧐
- Climate / Analyse Argo data 🧐
- Climate / Pangeo Notebook in Galaxy - Introduction to Xarray 🧐
- Sequence analysis / Clean and manage Sanger sequences from raw files to aligned consensus 🧐
- Sequence analysis / Mapping ✍️ 🧐
- Sequence analysis / Quality Control 🧐
- Sequence analysis / NCBI BLAST+ against the MAdLand 🧐
- Sequence analysis / Screening assembled genomes for contamination using NCBI FCS 📝 🧐
- Sequence analysis / SARS-CoV-2 Viral Sample Alignment and Variant Visualization 🧐
- Imaging / Object tracking using CellProfiler 🧐
- Imaging / Introduction to Image Analysis using Galaxy 🧐
- Imaging / Tracking of mitochondria and capturing mitoflashes 🧐
- Imaging / End-to-End Tissue Microarray Image Analysis with Galaxy-ME 🧐
- Imaging / Nucleoli segmentation and feature extraction using CellProfiler 🧐
- Imaging / Analyse HeLa fluorescence siRNA screen 🧐
- Statistics and machine learning / Deep Learning (Part 3) - Convolutional neural networks (CNN) 🧐
- Statistics and machine learning / Deep Learning (Part 1) - Feedforward neural networks (FNN) 🧐
- Statistics and machine learning / Interval-Wise Testing for omics data 🧐
- Statistics and machine learning / Text-mining with the SimText toolset 🧐
- Statistics and machine learning / PAPAA PI3K_OG: PanCancer Aberrant Pathway Activity Analysis 🧐
- Statistics and machine learning / Image classification in Galaxy with fruit 360 dataset 🧐
- Statistics and machine learning / Regression in Machine Learning 🧐
- Statistics and machine learning / Basics of machine learning 🧐
- Statistics and machine learning / Introduction to Machine Learning using R 🧐
- Statistics and machine learning / Fine tune large protein model (ProtTrans) using HuggingFace 🧐
- Statistics and machine learning / Supervised Learning with Hyperdimensional Computing 🧐
- Statistics and machine learning / Deep Learning (Part 2) - Recurrent neural networks (RNN) 🧐
- Statistics and machine learning / Machine learning: classification and regression 🧐
- Statistics and machine learning / A Docker-based interactive Jupyterlab powered by GPU for artificial intelligence in Galaxy 🧐
- Statistics and machine learning / Classification in Machine Learning 🧐
- Evolution / Tree thinking for tuberculosis evolution and epidemiology 🧐
- Evolution / Identifying tuberculosis transmission links: from SNPs to transmission clusters 🧐
- Epigenetics / Infinium Human Methylation BeadChip 🧐
- Epigenetics / Identification of the binding sites of the T-cell acute lymphocytic leukemia protein 1 (TAL1) 🧐
- Epigenetics / ATAC-Seq data analysis 🧐
- Epigenetics / CUT&RUN data analysis 📝 🧐
- Epigenetics / Formation of the Super-Structures on the Inactive X 🧐
- Epigenetics / Identification of the binding sites of the Estrogen receptor 🧐
- Epigenetics / DNA Methylation data analysis 🧐
- Epigenetics / Hi-C analysis of Drosophila melanogaster cells using HiCExplorer 🧐
- Proteomics / Proteogenomics 2: Database Search 🧐
- Proteomics / MaxQuant and MSstats for the analysis of label-free data 🧐
- Proteomics / Proteogenomics 3: Novel peptide analysis 🧐
- Proteomics / metaQuantome 1: Data creation 🧐
- Proteomics / Detection and quantitation of N-termini (degradomics) via N-TAILS 🧐
- Proteomics / Label-free versus Labelled - How to Choose Your Quantitation Method 🧐
- Proteomics / Protein FASTA Database Handling 🧐
- Proteomics / Clinical Metaproteomics 2: Discovery 🧐
- Proteomics / MaxQuant and MSstats for the analysis of TMT data 🧐
- Proteomics / Clinical Metaproteiomics 1: Database-Generation 🧐
- Proteomics / Secretome Prediction 🧐
- Proteomics / Machine Learning Modeling of Anticancer Peptides 🧐
- Proteomics / Metaproteomics tutorial 🧐
- Proteomics / Peptide and Protein ID using SearchGUI and PeptideShaker 🧐
- Proteomics / Proteogenomics 1: Database Creation 🧐
- Proteomics / EncyclopeDIA 🧐
- Proteomics / Peptide and Protein Quantification via Stable Isotope Labelling (SIL) 🧐
- Proteomics / Mass spectrometry imaging: Loading and exploring MSI data 🧐
- Proteomics / Clinical Metaproteomics 4: Quantitation 🧐
- Proteomics / Clinical Metaproteomics 3: Verification 🧐
- Proteomics / Peptide and Protein ID using OpenMS tools 🧐
- Proteomics / Clinical Metaproteomics 5: Data Interpretation 🧐
- Proteomics / Peptide Library Data Analysis 🧐
- FAIR Data, Workflows, and Research / Exporting Workflow Run RO-Crates from Galaxy 🧐
- FAIR Data, Workflows, and Research / FAIR in a nutshell 📝 🧐
- FAIR Data, Workflows, and Research / Metadata 🧐
- FAIR Data, Workflows, and Research / RO-Crate - Introduction 🧐
- FAIR Data, Workflows, and Research / Data Registration 🧐
- FAIR Data, Workflows, and Research / Best practices for workflows in GitHub repositories 🧐
- FAIR Data, Workflows, and Research / RO-Crate in Python 📝 🧐
- FAIR Data, Workflows, and Research / Introduction to Data Management Plan (DMP) for Peatland Research and PeatDataHub 📝 🧐
- FAIR Data, Workflows, and Research / Submitting workflows to LifeMonitor 📝 🧐
- FAIR Data, Workflows, and Research / Sequence data submission to ENA 🧐
- FAIR Data, Workflows, and Research / FAIR data management solutions 📝 🧐
- FAIR Data, Workflows, and Research / FAIRification of an RNAseq dataset 🧐
- FAIR Data, Workflows, and Research / Access 🧐
- FAIR Data, Workflows, and Research / REMBI - Recommended Metadata for Biological Images – metadata guidelines for bioimaging data 🧐
- FAIR Data, Workflows, and Research / FAIR Galaxy Training Material 📝 🧐
- FAIR Data, Workflows, and Research / Making clinical datasets FAIR 🧐
- FAIR Data, Workflows, and Research / FAIR Bioimage Metadata 🧐
- FAIR Data, Workflows, and Research / FAIR and its Origins 🧐
- FAIR Data, Workflows, and Research / Persistent Identifiers 🧐
- Contributing to the Galaxy Training Material / Creating Interactive Galaxy Tours 🧐
- Contributing to the Galaxy Training Material / Including a new topic 📝 🧐
- Contributing to the Galaxy Training Material / Teaching Python ✍️ 🧐
- Contributing to the Galaxy Training Material / GTN Metadata ✍️ 🧐
- Contributing to the Galaxy Training Material / Tools, Data, and Workflows for tutorials ✍️ 🧐
- Contributing to the Galaxy Training Material / Contributing with GitHub via command-line 🧐
- Contributing to the Galaxy Training Material / Creating content in Markdown ✍️ 🧐
- Contributing to the Galaxy Training Material / Creating a new tutorial ✍️ 🧐
- Contributing to the Galaxy Training Material / Running the GTN website locally using the command line 🧐
- Contributing to the Galaxy Training Material / Running the GTN website online using GitHub CodeSpaces 🧐
- Contributing to the Galaxy Training Material / Adding Quizzes to your Tutorial ✍️
- Contributing to the Galaxy Training Material / Adding auto-generated video to your slides ✍️ 🧐
- Contributing to the Galaxy Training Material / Principles of learning and how they apply to training and teaching 🧐
- Contributing to the Galaxy Training Material / Design and plan session, course, materials 🧐
- Contributing to the Galaxy Training Material / Single Cell Publication - Data Plotting ✍️
- Contributing to the Galaxy Training Material / Running the GTN website online using GitPod 🧐
- Contributing to the Galaxy Training Material / Single Cell Publication - Data Analysis ✍️
- Contributing to the Galaxy Training Material / FAIR-by-Design methodology 🧐
- Contributing to the Galaxy Training Material / Contributing with GitHub via its interface 🧐
- Contributing to the Galaxy Training Material / Updating diffs in admin training ✍️ 🧐
- Contributing to the Galaxy Training Material / Generating PDF artefacts of the website 🧐
- Foundations of Data Science / Advanced CLI in Galaxy ✍️ 🧐
- Foundations of Data Science / Advanced R in Galaxy 🧐
- Foundations of Data Science / Conda Environments For Software Development ✍️ 🧐
- Foundations of Data Science / SQL with R ✍️ 🧐
- Foundations of Data Science / Python - Type annotations ✍️ 🧐
- Foundations of Data Science / Advanced SQL ✍️ 🧐
- Foundations of Data Science / Python - Lists & Strings & Dictionaries ✍️ 🧐
- Foundations of Data Science / Make & Snakemake ✍️ 🧐
- Foundations of Data Science / Virtual Environments For Software Development ✍️ 🧐
- Foundations of Data Science / Python - Testing ✍️ 🧐
- Foundations of Data Science / Introduction to Python 🧐
- Foundations of Data Science / Data Manipulation Olympics - JQ ✍️
- Foundations of Data Science / dplyr & tidyverse for data processing ✍️ 🧐
- Foundations of Data Science / Python - Multiprocessing ✍️ 🧐
- Foundations of Data Science / SQL Educational Game - Murder Mystery ✍️ 🧐
- Foundations of Data Science / Advanced Python 🧐
- Foundations of Data Science / Python - Coding Style 📝
- Foundations of Data Science / Python - Try & Except ✍️ 🧐
- Foundations of Data Science / Python - Argparse ✍️ 🧐
- Foundations of Data Science / R basics in Galaxy 🧐
- Foundations of Data Science / Introduction to sequencing with Python (part one) 🧐
- Foundations of Data Science / Python - Functions ✍️ 🧐
- Foundations of Data Science / Python - Files & CSV ✍️ 🧐
- Foundations of Data Science / Version Control with Git 📝 🧐
- Foundations of Data Science / Variant Calling Workflow ✍️ 🧐
- Foundations of Data Science / CLI basics ✍️ 🧐
- Foundations of Data Science / A (very) brief history of genomics 🧐
- Foundations of Data Science / Python - Globbing ✍️ 🧐
- Foundations of Data Science / Basics of using Git from the Command Line ✍️ 🧐
- Foundations of Data Science / CLI Educational Game - Bashcrawl ✍️ 🧐
- Foundations of Data Science / Plotting in Python 🧐
- Foundations of Data Science / Python - Math ✍️ 🧐
- Foundations of Data Science / Introduction to SQL ✍️ 🧐
- Foundations of Data Science / Data Manipulation Olympics - SQL ✍️
- Foundations of Data Science / Data visualisation Olympics - Visualization in R 📝
- Foundations of Data Science / Python - Loops ✍️ 🧐
- Foundations of Data Science / Python - Flow Control ✍️ 🧐
- Foundations of Data Science / Python - Basic Types & Type Conversion ✍️ 🧐
- Foundations of Data Science / SQL with Python ✍️ 🧐
- Foundations of Data Science / Python - Subprocess ✍️ 🧐
- Foundations of Data Science / Python - Introductory Graduation ✍️ 🧐
- Transcriptomics / Pathway analysis with the MINERVA Platform ✍️ ⚙️ 🧐
- Transcriptomics / Genome-wide alternative splicing analysis 🧐
- Transcriptomics / Differential abundance testing of small RNAs 🧐
- Transcriptomics / 3: RNA-seq genes to pathways 🧐
- Transcriptomics / De novo transcriptome reconstruction with RNA-Seq 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis 📝 🧐
- Transcriptomics / Visualization of RNA-Seq results with Volcano Plot 🧐
- Transcriptomics / CLIP-Seq data analysis from pre-processing to motif detection 🧐
- Transcriptomics / 1: RNA-Seq reads to counts 🧐
- Transcriptomics / RNA-Seq analysis with AskOmics Interactive Tool 🧐
- Transcriptomics / Small Non-coding RNA Clustering using BlockClust 🧐
- Transcriptomics / 2: RNA-seq counts to genes 🧐
- Transcriptomics / Visualization of RNA-Seq results with CummeRbund 🧐
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana 🧐
- Transcriptomics / GO Enrichment Analysis 🧐
- Transcriptomics / Reference-based RNAseq data analysis (long) 🧐
- Transcriptomics / RNA Seq Counts to Viz in R 🧐
- Transcriptomics / Network analysis with Heinz 🧐
- Transcriptomics / Visualization of RNA-Seq results with heatmap2 🧐
- Transcriptomics / RNA-seq Alignment with STAR 📝 🧐
- Variant Analysis / Calling very rare variants 🧐
- Variant Analysis / Identification of somatic and germline variants from tumor and normal sample pairs 🧐
- Variant Analysis / Calling variants in non-diploid systems 🧐
- Variant Analysis / Pox virus genome analysis from tiled-amplicon sequencing data 🧐
- Variant Analysis / Calling variants in diploid systems 🧐
- Variant Analysis / Mapping and molecular identification of phenotype-causing mutations 🧐
- Variant Analysis / Exome sequencing data analysis for diagnosing a genetic disease 🧐
- Variant Analysis / Trio Analysis using Synthetic Datasets from RD-Connect GPAP ✍️ 🧐
- Variant Analysis / From NCBI's Sequence Read Archive (SRA) to Galaxy: SARS-CoV-2 variant analysis 🧐
- Variant Analysis / Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data 🧐
- Variant Analysis / M. tuberculosis Variant Analysis 🧐
- Variant Analysis / Somatic Variant Discovery from WES Data Using Control-FREEC 🧐
- Variant Analysis / Microbial Variant Calling 🧐
- Variant Analysis / Avian influenza viral strain analysis from gene segment sequencing data 🧐
- Genome Annotation / Long non-coding RNAs (lncRNAs) annotation with FEELnc 🧐
- Genome Annotation / Functional annotation of protein sequences 🧐
- Genome Annotation / Refining Genome Annotations with Apollo (prokaryotes) ✍️ 🧐
- Genome Annotation / Comparative gene analysis in unannotated genomes 🧐
- Genome Annotation / Masking repeats with RepeatMasker 🧐
- Genome Annotation / Identification of AMR genes in an assembled bacterial genome 📝 🧐
- Genome Annotation / From small to large-scale genome comparison 🧐
- Genome Annotation / Genome annotation with Funannotate 🧐
- Genome Annotation / Genome annotation with Maker 🧐
- Genome Annotation / Genome annotation with Helixer 🧐
- Genome Annotation / Genome annotation with Maker (short) 🧐
- Genome Annotation / Genome annotation with Prokka 🧐
- Genome Annotation / CRISPR screen analysis 🧐
- Genome Annotation / Genome Annotation 🧐
- Genome Annotation / Creating an Official Gene Set 🧐
- Genome Annotation / Bacterial Genome Annotation 🧐
- Genome Annotation / Essential genes detection with Transposon insertion sequencing 🧐
- Genome Annotation / Refining Genome Annotations with Apollo (eukaryotes) 🧐
- Development in Galaxy / Scripting Galaxy using the API and BioBlend 📝 🧐
- Development in Galaxy / Data source integration ✍️ 🧐
- Development in Galaxy / Adding and updating best practice metadata for Galaxy tools using the bio.tools registry 🧐
- Development in Galaxy / ToolFactory: Generating Tools From Simple Scripts ✍️ 🧐
- Development in Galaxy / JavaScript plugins 🧐
- Development in Galaxy / Galaxy Interactive Tools ✍️ 🧐
- Development in Galaxy / Debugging Galaxy 🧐
- Development in Galaxy / Generic plugins 🧐
- Development in Galaxy / ToolFactory: Generating Tools From More Complex Scripts 🧐
- Development in Galaxy / Creating Galaxy tools from Conda Through Deployment 🧐
- Development in Galaxy / Galaxy Webhooks 🧐
- Development in Galaxy / Contributing a New Feature to Galaxy Core 🧐
- Development in Galaxy / Contributing to BioBlend as a developer 🧐
- Development in Galaxy / Writing Automated Tests for Galaxy 🧐
- Materials Science / Finding the muon stopping site with pymuon-suite in Galaxy 🧐
- Introduction to Galaxy Analyses / Very Short Introductions: QC 🧐
- Introduction to Galaxy Analyses / Introduction to Genomics and Galaxy 🧐
- Introduction to Galaxy Analyses / A short introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / From peaks to genes ✍️ 🧐
- Introduction to Galaxy Analyses / IGV Introduction 🧐
- Introduction to Galaxy Analyses / NGS data logistics 🧐
- Introduction to Galaxy Analyses / Upload data to Galaxy 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for genomics ✍️ 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for everyone 🧐
- Introduction to Galaxy Analyses / How to reproduce published Galaxy analyses 🧐
- Introduction to Galaxy Analyses / Data Manipulation Olympics 📝 🧐
- Galaxy Community Building / Creation of an interactive Galaxy tools table for your community 🧐
- Galaxy Community Building / Make your tools available on your subdomain 🧐
- Galaxy Community Building / What's a Special Interest Group? 🧐
- Galaxy Community Building / Creating a Special Interest Group 🧐
- Galaxy Community Building / Creating community content 🧐
- Metabolomics / Mass spectrometry: GC-MS data processing (with XCMS, RAMClustR, RIAssigner, and matchms) 📝 🧐
- Metabolomics / Mass spectrometry: LC-MS preprocessing with XCMS 🧐
- Metabolomics / Mass spectrometry imaging: Examining the spatial distribution of analytes 🧐
- Metabolomics / Mass spectrometry: LC-MS data processing 🧐
- Metabolomics / Mass spectrometry: LC-MS analysis 🧐
- Single Cell / Filtrado, representación y exploración de secuenciación de ARN de células únicas 🧐
- Single Cell / Generación de una matriz de datos de secuenciación de ARN de células únicas utilizando Alevin 🧐
- Single Cell / Generación de una matriz de datos de secuenciación de ARN de células únicas utilizando Alevin 🧐
- Introduction to Galaxy Analyses / Breve introducción a Galaxy - en español 🧐
- Ecology / Production d'indicateurs champs de bloc 🧐
Slides
- Synthetic Biology / Introduction to Synthetic Biology 🧐
- Development in Galaxy / Galaxy from a developer point of view 🧐
- Materials Science / Introduction to Muon Spectroscopy 🧐
- Microbiome / Introduction to metatranscriptomics 🧐
- Microbiome / Introduction to Microbiome Analysis 🧐
- Using Galaxy and Managing your Data / Galaxy workflows in Dockstore 🧐
- Using Galaxy and Managing your Data / Introduction to SRA Aligned Read Format and Cloud Metadata for SARS-CoV-2 🧐
- Using Galaxy and Managing your Data / Getting data into Galaxy 🧐
- Galaxy Server administration / Terraform ✍️ 🧐
- Galaxy Server administration / Controlling Galaxy with systemd or Supervisor ✍️ 🧐
- Galaxy Server administration / uWSGI 🧐
- Galaxy Server administration / Galaxy Installation with Ansible 📝 🧐
- Galaxy Server administration / Galaxy Monitoring ✍️
- Galaxy Server administration / Gearing towards production 🧐
- Galaxy Server administration / Server Maintenance: Cleanup, Backup, and Restoration ✍️ 🧐
- Galaxy Server administration / Galaxy Interactive Tools 🧐
- Galaxy Server administration / Reference Data with CVMFS ✍️ 🧐
- Galaxy Server administration / Galaxy Administrator Time Burden and Technology Usage 📝 🧐
- Galaxy Server administration / Storage Management ✍️ 🧐
- Galaxy Server administration / Galaxy on the Cloud 📝 🧐
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar ✍️ 🧐
- Galaxy Server administration / Ansible ✍️ 🧐
- Galaxy Server administration / Reference Genomes in Galaxy 🧐
- Galaxy Server administration / Galaxy Monitoring with gxadmin ✍️ 🧐
- Galaxy Server administration / External Authentication 🧐
- Galaxy Server administration / Empathy ✍️ 🧐
- Galaxy Server administration / Advanced customisation of a Galaxy instance ✍️ 🧐
- Galaxy Server administration / Storage Management ✍️ 🧐
- Galaxy Server administration / Galaxy and Celery 📝 🧐
- Galaxy Server administration / Galactic Database ✍️ 🧐
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana ✍️ 🧐
- Galaxy Server administration / User, Role, Group, Quota, and Authentication managment ✍️ 🧐
- Galaxy Server administration / Galaxy Tool Management with Ephemeris ✍️ 🧐
- Galaxy Server administration / Galaxy Troubleshooting 🧐
- Galaxy Server administration / Galaxy from an administrator's point of view ✍️ 🧐
- Galaxy Server administration / Connecting Galaxy to a compute cluster ✍️ 🧐
- Galaxy Server administration / Docker and Galaxy 🧐
- Galaxy Server administration / Server: Other ✍️ 🧐
- Visualisation / Friends Don't Let Friends Make Bad Graphs 📝
- Visualisation / JBrowse ✍️ 🧐
- Visualisation / Visualisations in Galaxy ✍️ 🧐
- Visualisation / Circos ✍️ 🧐
- Single Cell / Trajectory analysis 🧐
- Single Cell / Plates, Batches, and Barcodes 🧐
- Single Cell / An introduction to scRNA-seq data analysis 🧐
- Single Cell / Dealing with Cross-Contamination in Fixed Barcode Protocols 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy 🧐
- Single Cell / Automated Cell Annotation 🧐
- Teaching and Hosting Galaxy training / Overview of the Galaxy Training Material for Instructors ✍️
- Teaching and Hosting Galaxy training / Workshop Kickoff 🧐
- Assembly / De Bruijn Graph Assembly 🧐
- Assembly / An introduction to get started in genome assembly and annotation 🧐
- Assembly / Genome assembly quality control. 🧐
- Assembly / Deeper look into Genome Assembly algorithms 🧐
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads 🧐
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / Unicycler Assembly 🧐
- Climate / Pangeo ecosystem 101 for everyone 🧐
- Climate / Functionally Assembled Terrestrial Ecosystem Simulator (FATES) 🧐
- Climate / Introduction to climate data 🧐
- Climate / The Pangeo ecosystem 🧐
- Sequence analysis / Mapping 🧐
- Sequence analysis / Quality Control 🧐
- Statistics and machine learning / Convolutional neural networks (CNN) Deep Learning - Part 3 🧐
- Statistics and machine learning / Feedforward neural networks (FNN) Deep Learning - Part 1 🧐
- Statistics and machine learning / Image classification in Galaxy with fruit 360 dataset 🧐
- Evolution / Phylogenetics - Back to Basics - Estimating trees from alignments 🧐
- Evolution / Phylogenetics - Back to Basics - Terminology 🧐
- Evolution / Phylogenetics - Back to Basics - Building Trees 🧐
- Evolution / Phylogenetics - Back to Basics - Phylogenetic Networks 🧐
- Evolution / Phylogenetics - Back to Basics - Multiple Sequence Alignment 🧐
- Evolution / Phylogenetics - Back to Basics - Introduction 🧐
- Epigenetics / EWAS Epigenome-Wide Association Studies Introduction 🧐
- Epigenetics / Introduction to DNA Methylation data analysis 🧐
- Epigenetics / Introduction to ATAC-Seq data analysis 🧐
- Epigenetics / ChIP-seq data analysis 🧐
- Epigenetics / Introduction to ChIP-Seq data analysis 🧐
- Proteomics / Introduction to proteomics, protein identification, quantification and statistical modelling 🧐
- FAIR Data, Workflows, and Research / Intro to DataPLANT ARCs 🧐
- Contributing to the Galaxy Training Material / Contributing with GitHub via command-line 🧐
- Contributing to the Galaxy Training Material / Creating Slides ✍️ 🧐
- Contributing to the Galaxy Training Material / Overview of the Galaxy Training Material 🧐
- Foundations of Data Science / A brief history of modern biology 🧐
- Transcriptomics / Identification of non-canonical ORFs and their potential biological function 🧐
- Transcriptomics / Integrate and query local datasets and distant RDF data with AskOmics using Semantic Web technologies 🧐
- Transcriptomics / Visualization of RNA-Seq results with CummeRbund 🧐
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana 🧐
- Transcriptomics / Network Analysis with Heinz 🧐
- Transcriptomics / Introduction to Transcriptomics 🧐
- Genome Annotation / Refining Genome Annotations with Apollo ✍️ 🧐
- Genome Annotation / High Performance Computing for Pairwise Genome Comparison 🧐
- Genome Annotation / Genome annotation with Prokka 🧐
- Genome Annotation / Introduction to CRISPR screen analysis 🧐
- Genome Annotation / Introduction to Genome Annotation ✍️ 🧐
- Development in Galaxy / Scripting Galaxy using the API and BioBlend 🧐
- Development in Galaxy / Galaxy Interactive Tours 🧐
- Development in Galaxy / Tool Shed: sharing Galaxy tools 🧐
- Development in Galaxy / Introduction to the ToolFactory tutorial. 🧐
- Development in Galaxy / Visualizations: JavaScript Plugins 🧐
- Development in Galaxy / Generic plugins 🧐
- Development in Galaxy / Tool Dependencies and Containers 🧐
- Development in Galaxy / Tool Dependencies and Conda 🧐
- Development in Galaxy / Galaxy Interactive Environments ✍️ 🧐
- Development in Galaxy / Tool development and integration into Galaxy 🧐
- Development in Galaxy / Galaxy Webhooks 🧐
- Development in Galaxy / Prerequisites for building software/conda packages 🧐
- Development in Galaxy / Galaxy Code Architecture 🧐
- Introduction to Galaxy Analyses / A Short Introduction to Galaxy ✍️ 🧐
- Introduction to Galaxy Analyses / Options for using Galaxy 🧐
- Introduction to Galaxy Analyses / Introduction to Galaxy ✍️ 🧐
- Metabolomics / Introduction to Metabolomics 🧐
- Single Cell / Introducción al análisis de datos de scRNA-seq 🧐
- Introduction to Galaxy Analyses / Una breve introducción a Galaxy ✍️ 🧐
- Single Cell / Una introducción al análisis de datos scRNA-seq 🧐
- Introduction to Galaxy Analyses / Una Breve Introducción a Galaxy ✍️ 🧐
FAQs
- How does the GTN ensure accessibility?
- Operating system compatibility
- Beware of Cuts
- Will my jobs keep running?
- What information should I include when reporting a problem?
- Variable connection
- What if you forget `--diff`?
- Error: "skipping: no hosts matched"
- Running Ansible on your remote machine
- What is the difference between the roles with `role:` prefix and without?
- How do I know what I can do with a role? What variables are available?
- How do I see what variables are set for a host?
- Is YAML sensitive to True/true/False/false
- Opening a split screen in byobu
- Can I use a public Galaxy for my private data?
- How does the GTN implement the "Ten simple rules for collaborative lesson development"
- Adding a tag to a collection
- Creating a dataset collection
- Creating a paired collection
- Changing the datatype of a collection
- Renaming a collection
- How do I find the Community Home pages?
- How to Contribute to Galaxy
- Contributing a Jupyter Notebook to the GTN
- How can I contribute in "advanced" mode?
- Customising the welcome page
- Adding a tag
- Changing the datatype
- Changing database/build (dbkey)
- Converting the file format
- Creating a new file
- Detecting the datatype (file format)
- Importing data from a data library
- Importing data from remote files
- Importing via links
- Renaming a dataset
- How to read a Diff
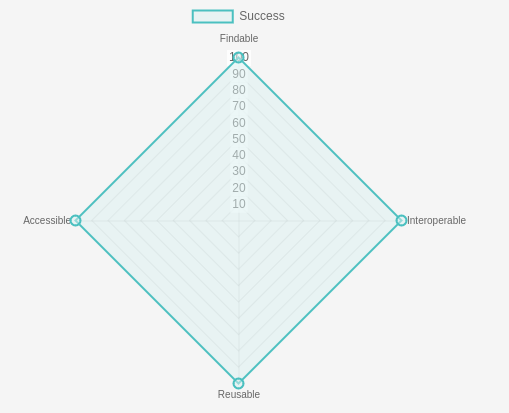
- How does the GTN ensure our training materials are FAIR?
- Using the Window Manager to view multiple datasets
- Flatten a list of list of paired datasets into a list of paired datasets
- How many mules?
- How can I get my container requiring jobs to run in a container?
- Updating from 22.01 to 23.0 with Ansible
- Compatible Versions of Galaxy
- Using Git With Ansible Vaults
- Time to git commit
- Galaxy Admin Training Path
- Forking the GTN repository
- Updating the default branch from master to main
- Syncing your Fork of the GTN
- GTN ADR: Image Storage
- GTN ADR: Why Jekyll and not another Static Site Generator (SSG)
- GTN Architectural Decision Record Template
- What is an Architectural Decision Record (ADR)?
- Slow incremental builds
- Creating a GTN FAQ
- What licenses are used in the GTN?
- GTN Stats
- Adding workflow tests with Planemo
- Copy a dataset between histories
- Créer un nouvel history
- Creating a new history
- View a list of all histories
- Renaming a history
- Searching your history
- Input Histories & Answer Keys
- Open a Terminal in Jupyter
- Knitting RMarkdown documents in RStudio
- Launch RStudio
- Learning with RMarkdown in RStudio
- TB Variant Report crashes (with an error about KeyError: 'protein')
- Library Permission Issues
- Failing all jobs from a specific user
- How do I find the Maintainer Home pages?
- Mapping Jobs to Specific Storage By User
- How do I add a news feed to a Matrix channel?
- Got lost along the way?
- What is my.galaxy.training
- Using the new Contributions Annotation framework
- Getting your API key
- Debugging Memory Leaks
- Quality Scores
- Recording a video tutorial
- Preparing materials for asynchronous learning: CYOA
- Preparing materials for asynchronous learning: FAQs
- Preparing materials for asynchronous learning: Self-Study
- Preparing materials for asynchronous learning: Tips
- Making an element collapsible in a report
- Enhancing tabular dataset previews in reports/pages
- Finding a material's PURL or Short URL
- Re-running a tool
- Selecting a dataset collection as input
- Select multiple datasets
- Translations within the GTN
- Add genome and annotations to IGV from Galaxy
- Why isn't my history updating?
- Annotating Pre-requisites
- Why host your materials with the GTN?
- Why not use Excel?
- Ensuring Workflows meet Best Practices
- Creating a new workflow
- Opening the workflow editor
- Extracting a workflow from your history
- Hiding intermediate steps
- Importing a workflow
- Setting parameters at run-time
- Renaming workflow outputs
- Viewing a workflow report
- Running a workflow
- Importing and Launching a Dockstore Workflow
- Importing and launching a GTN workflow
- Importing and Launching a WorkflowHub.eu Workflow
Video Recordings
- Galaxy Server administration / Reference Data with CVMFS 💬
- Galaxy Server administration / Galaxy Monitoring with gxadmin 💬
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana 💬
- Galaxy Server administration / User, Role, Group, Quota, and Authentication managment 💬
- Visualisation / Circos 🗣
- Assembly / An Introduction to Genome Assembly 💬
- Sequence analysis / Quality Control 💬
- Galaxy Server administration / Galaxy Installation with Ansible 💬 🗣
- Galaxy Server administration / Galaxy Interactive Tools 💬
- Galaxy Server administration / Use Apptainer containers for running Galaxy jobs 💬 🗣
- Galaxy Server administration / Data Libraries 💬
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar 💬
- Galaxy Server administration / Performant Uploads with TUS 💬 🗣
- Galaxy Server administration / Mapping Jobs to Destinations using TPV 🗣
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana 💬
- Galaxy Server administration / Training Infrastructure as a Service (TIaaS) 💬 🗣
- Galaxy Server administration / Connecting Galaxy to a compute cluster 🗣
- Galaxy Server administration / Upgrading Galaxy 💬
- Visualisation / Visualisation with Circos 💬 🗣
- Transcriptomics / Reference-based RNA-Seq data analysis 💬
- Transcriptomics / RNA Seq Counts to Viz in R 💬
- Variant Analysis / Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data 💬
- Variant Analysis / M. tuberculosis Variant Analysis 💬
Events
- Admin Training @ GCC 2022: Online Training Day 🎪 🧑🏫
- 2023 Galaxy Admin Training (Ghent) 🧑🏫
- My Training Event Title 🎪
GitHub Activity
github Issues Reported
744 Merged Pull Requests
See all of the github Pull Requests and github Commits by Helena Rasche.
-
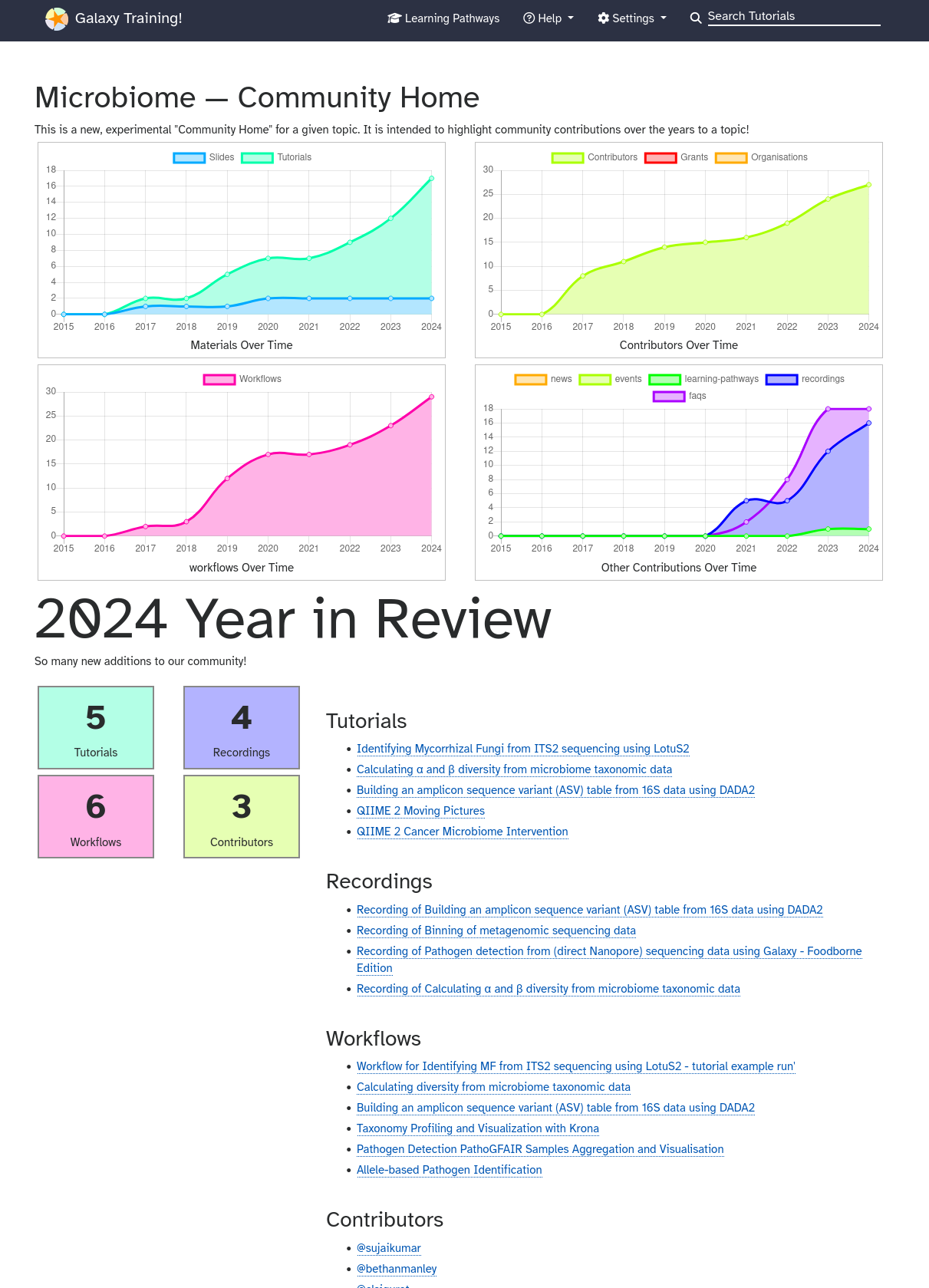
 GTN Year In Review
admindevvariant-analysisintroductiontranscriptomics
GTN Year In Review
admindevvariant-analysisintroductiontranscriptomics -
 code documentation
admintemplate-and-toolscontributingdata-science
code documentation
admintemplate-and-toolscontributingdata-science -
 Improve single cell topic page
template-and-toolssingle-cell
Improve single cell topic page
template-and-toolssingle-cell -
 improved by-tools page
introductiontemplate-and-toolsassemblyepigeneticsstatistics
improved by-tools page
introductiontemplate-and-toolsassemblyepigeneticsstatistics -
 Fix beacon tutorial issues
admin
Fix beacon tutorial issues
admin
Reviewed 1422 PRs
We love our community reviewing each other's work!
-
 Add some links to community home pages to Wendi's news posts
news
Add some links to community home pages to Wendi's news posts
news -
 Add borders
Add borders
-
 Add community content resource page
template-and-toolsnews
Add community content resource page
template-and-toolsnews -
 Update 2025-01-28-src.md
admintemplate-and-tools
Update 2025-01-28-src.md
admintemplate-and-tools -
 Add end-of-year blog post for SPOC Community
news
Add end-of-year blog post for SPOC Community
news
News

Next GTN CoFest May 20, 2021
18 March 2021
Every 3 months we organise one day dedicated to the GTN community! Thursday, May 20th is the next GTN CoFest. We will be working on the training materials, have discussions with the global Galaxy traning community. Do you teach with Galaxy? Want to learn how to add your tutorial to the GTN? New to the community and just want to learn more? Everybody is welcome!

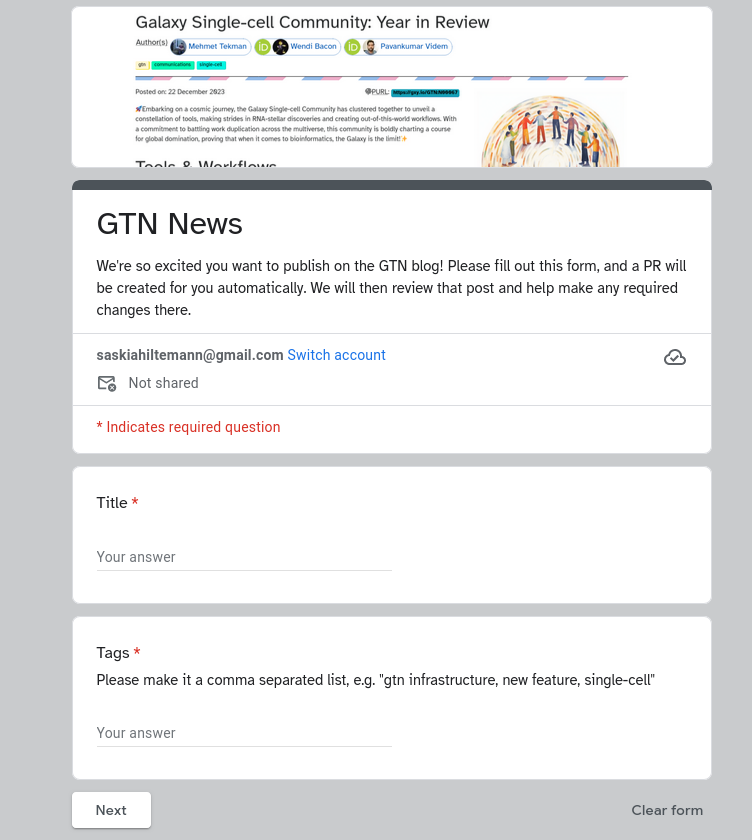
New Feature: FAQs
24 March 2021
Snippets have received an major overhaul! Snippets/FAQs are small reusable bits of training, often answering a single question. In addition to the general Galaxy snippets, we now also support topic-level and tutorial-level FAQs. This will allow contributors to add common questions and answers to their tutorials, which not only can be included in the tutorial text itself, but are also used to autogenerate an FAQ page for the tutorial or topic listing all common questions (and answers).This will be useful both to participants following tutorials via self-study, and also to instructors preparing for a training event.

New Tutorials: Genome assembly of a MRSA genome
24 March 2021
Two new assembly tutorials have been added to the GTN! These tutorials walk you through the
process of assembling MRSA strains from real-world datasets. MRSA stands for “Methicillin-
resistant Staphylococcus aureus, it is resistance to several antibiotics and is a common
source of hospital-acquired infections and outbreaks. In this tutorial you will assess the
quality of your data, perform an assembly and assess its quality, analyze your data for the
presence of antimicrobial resistance (AMR) genes, and annotate the genome using Prokka and
JBrowse.
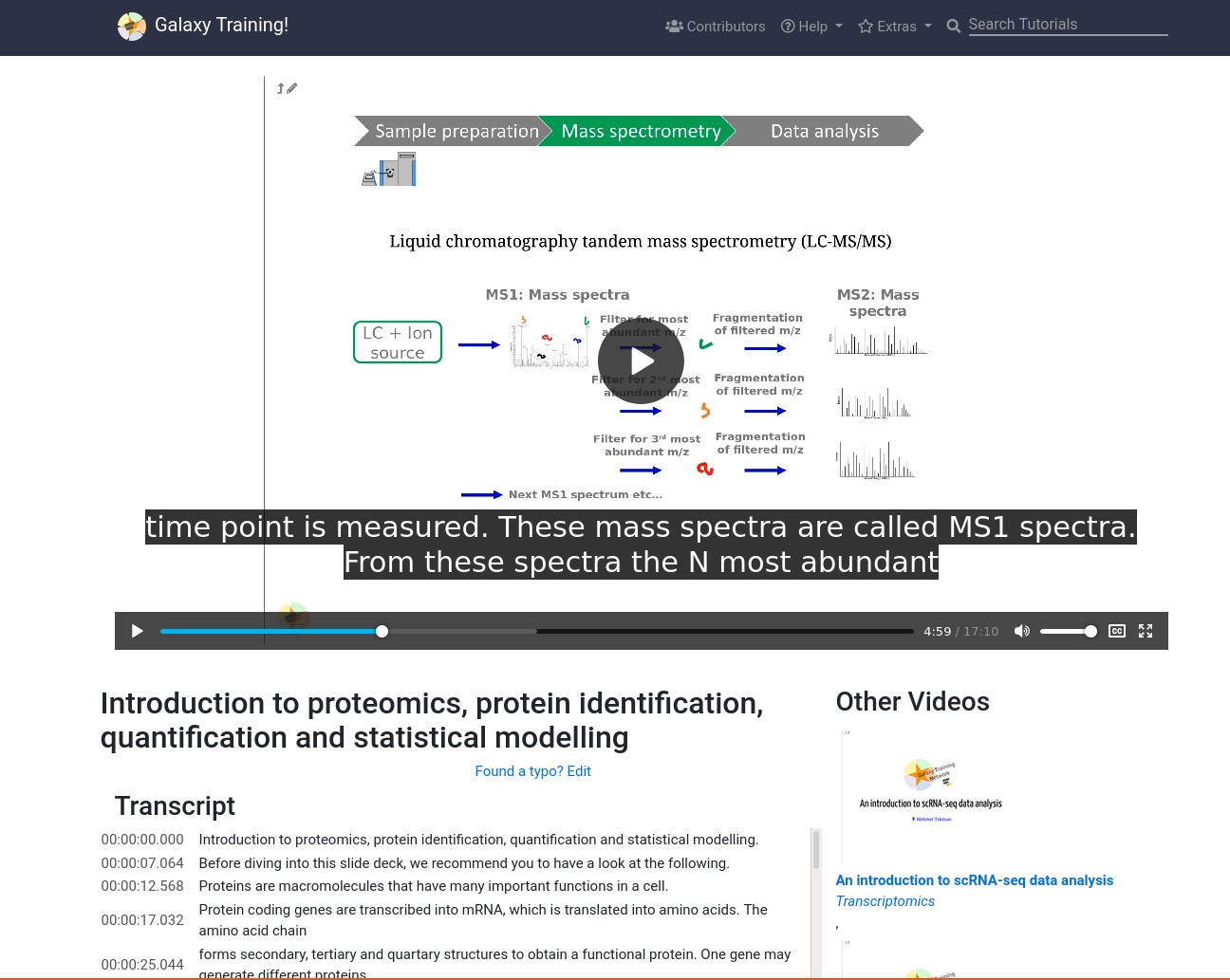
New Feature: Video Player
6 April 2021
One of our contributors (@gtn:mtekman) found himself wanting to link to a specific time point in a video. We said “Wow, what a fantastic idea!” and then we implemented it. To support that we developed a very simple standalone video player page where you can view the GTN videos and browse through our catalog. It features the full transcript on the left, some suggested videos on the right, and a very standard <video> player.

¿Hablas español?: The first curated tutorial in Spanish!
20 May 2021
¡El primer tutorial en español ya está disponible! Galaxy siempre ha tenido tutoriales traducidos automáticamente mediante Google-Translate, ahora nos estamos embarcando en un nuevo proyecto para estudiar la experiencia de aprendizaje con tutoriales en bioinformática traducidos por humanos. Nuestro objetivo es crear un paquete de tutoriales para el análisis de datos single-cell (célula única) en primera instancia, con el fin de estudiar su uso y utilidad en un taller para hablantes nativos de español. Agradecemos enormemente a las nuevas participantes @gtn:pclo y @gtn:ales-ibt (EMBL-EBI) así como a las miembros existentes de GTN @gtn:beatrizserrano (University of Freiburg), @gtn:shiltemann & @gtn:hexylena (Erasmus-MC), por su arduo trabajo en este proyecto de traducción y mantenimiento y, por supuesto, a @gtn:nomadscientist (The Open University/EBI) como líder de proyecto.

Oh no, it changed! Quick, to the archive menu.
1 June 2021
Did you ever start teaching a GTN tutorial, only to realise something changed recently? Oh no! A while ago the GTN created an Archive to address just this concern.
On every tag, we’d build a copy of the website which we would place online and never touch again. This way we’d have permanent online copies so that if someone needed to go back to an older version of the GTN, they could.
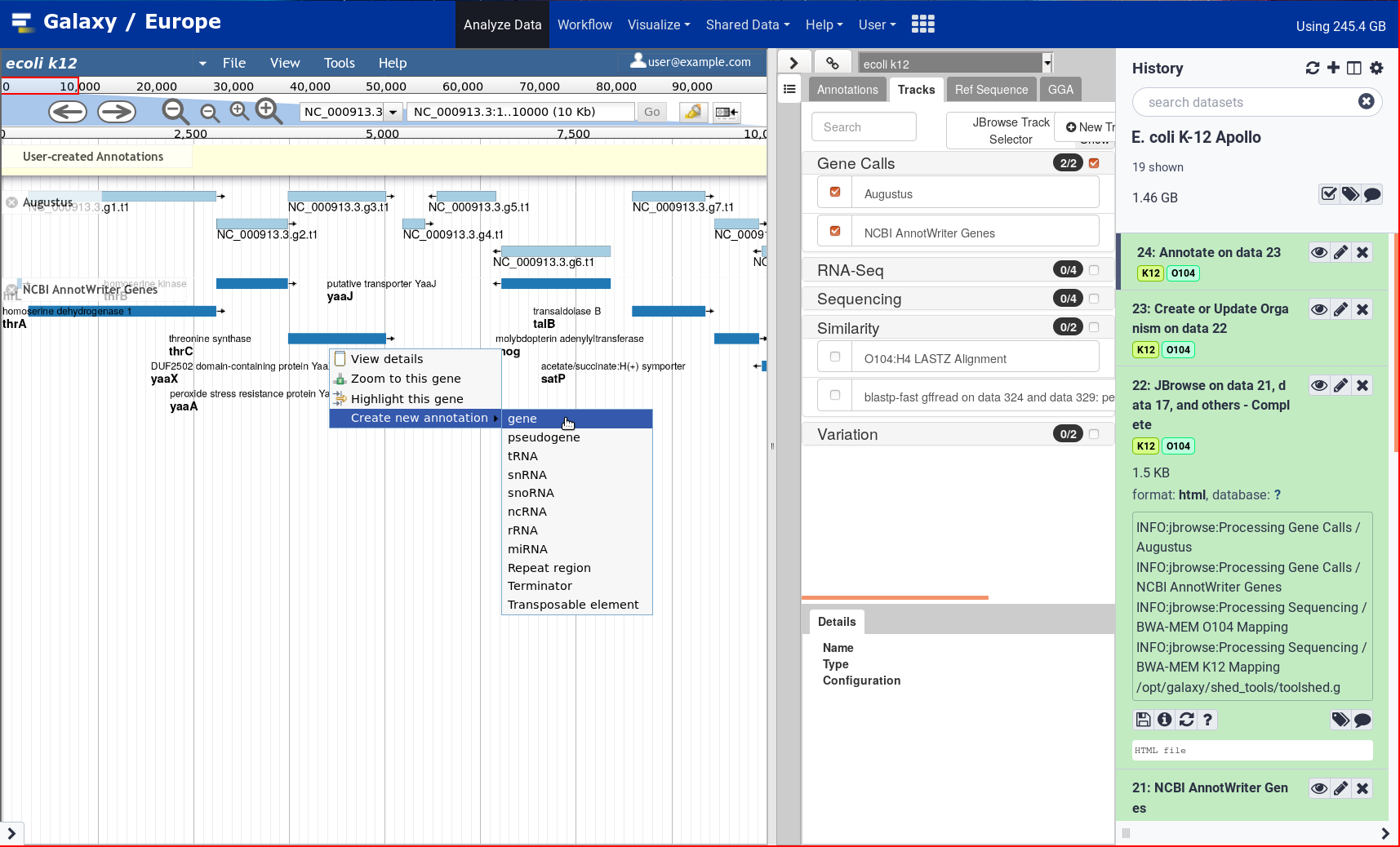
New Tutorial: Genome Annotation with Apollo
4 June 2021
After two years, the Galaxy Genome Annotation team has finally finished their much awaited tutorial on doing Genome Annotation in Galaxy! Genome annotation is a time consuming and largely manual process as it requires the synthesis of a significant amount of varied information to make inferences, a process tha cannot be accomplished without an interactive editor. To support this, Apollo was deployed on UseGalaxy.eu. Over the past two years the GGA project has pushed forward apollo and tooling around it, and it finally culminates in this look at how to annotated your genomes in Apollo.
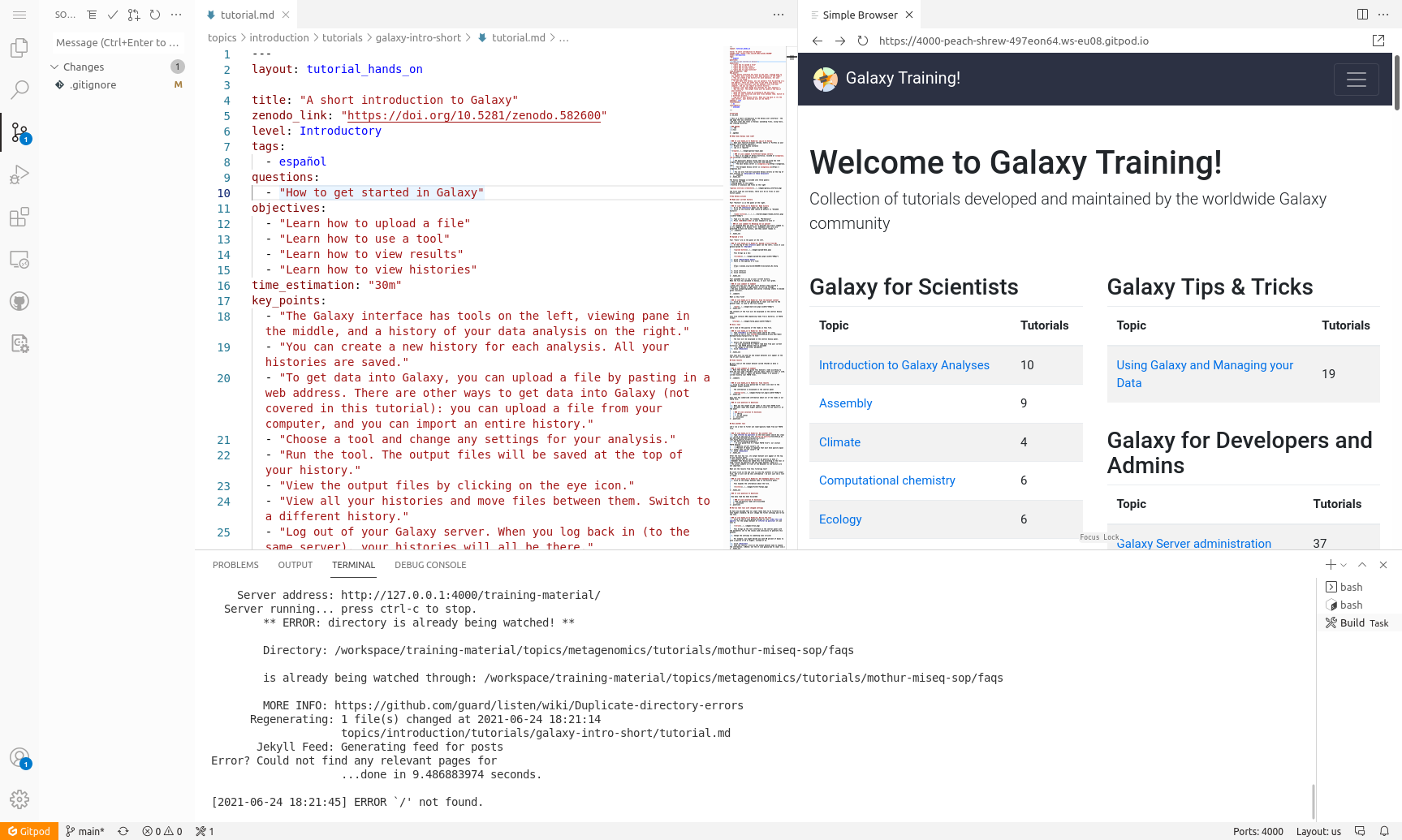
New Tutorial: GitPod for contributing to the GTN
25 June 2021
Want to contribute to the GTN without installing anything on your own computer? GitPod provides an online workspace for editing the GTN tutorials, and showing a live preview of the GTN website with your changes. This tutorial shows you how to set up GitPod, how to use it to get a live preview of the GTN website, and how to save any changes you make back to your GitHub fork. This makes it easier than ever to contribute to the GTN!

Accessibility Improvements
27 July 2021
One of our community members suggested on twitter that we support alternative formats for accessing our JavaScript based slides, as this can be difficult for users using screen readers. We’ve now added support for “plain text slides”, where we render the slide decks as a single long document.
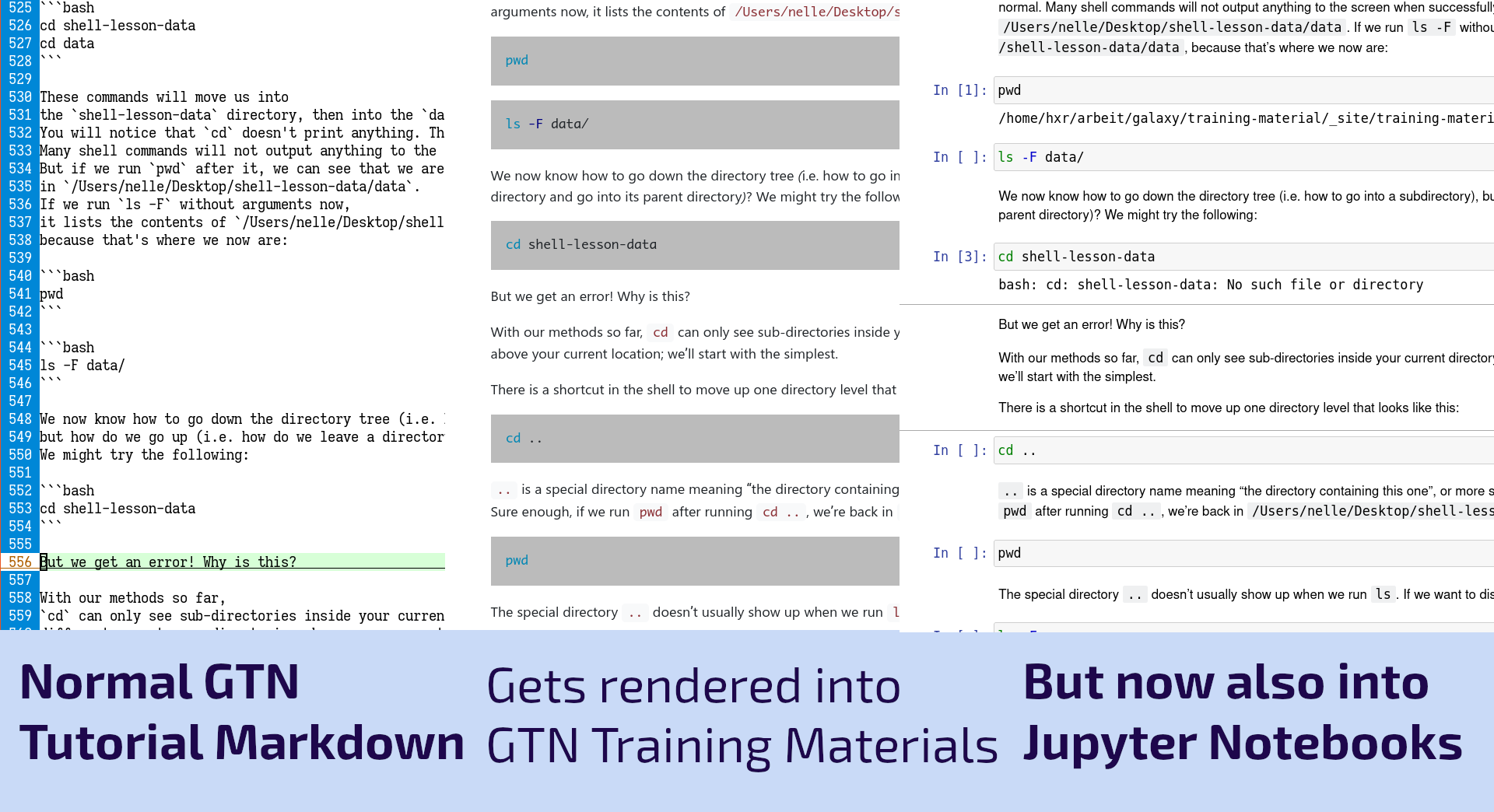
New Feature: Automatic Jupyter Notebooks
24 September 2021
As part of the work for the Gallantries Grant, we found ourselves wanting to teach a number of non-Galaxy tutorials on topics such as Python, Bash, SQL, and Git. Writing the tutorials was always a bit awkward as you would spend a lot of time writing code blocks that would be copied and pasted by students into a terminal. We thought we could do better than that, so we decided we would try to convert these to Jupyter Notebooks where the students could read the training materials and simultaneously execute the code cells directly there with the text! This worked fantastically and now we’ve built this feature into the GTN to make it easier to run all of the coding tutorials.
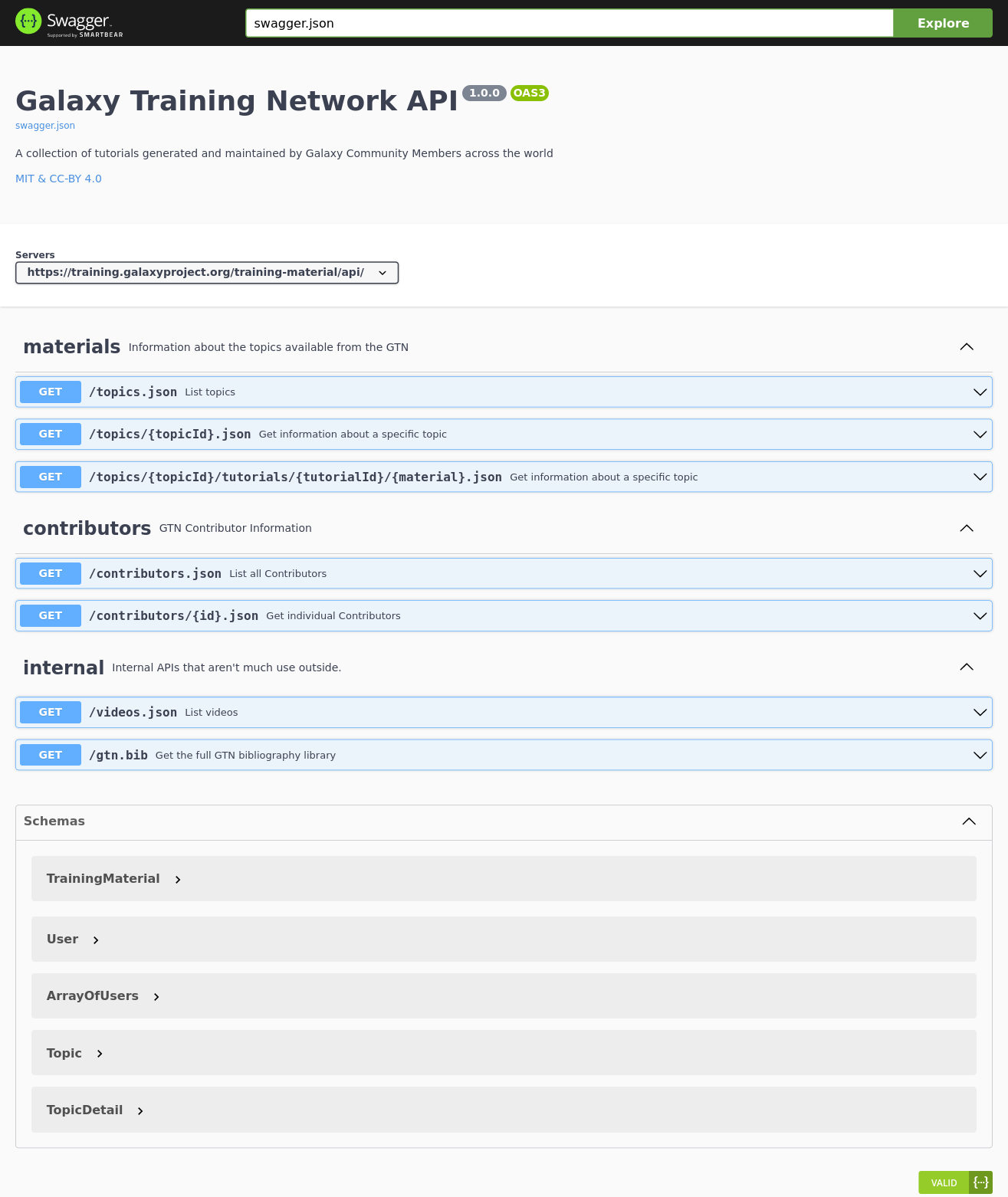
New Feature: GTN API with OpenAPI 3 specification
10 November 2021
As part of the work for the Gallantries Grant, we wanted to access some of the data from the GTN in a more accessible, API based way. We are working on building a Video Library based on the wild success of our Smörgåsbord and GCC2021 events and to support that we wanted to start pulling in data from the GTN to save course authors time and effort when designing their asynchronous courses.

New FAQs: How does the GTN stay FAIR and Collaborative
1 December 2021
The GTN does a lot of work to adhere to the best practices for FAIR training materials as set out in Garcia et al. 2020 and a lot of this work is behind-the-scenes or automatic from the infrastructure we provide, and we wanted to highlight that for educators and organisers who want to use GTN materials. This is one of the ways the GTN works to makes itself a fantastic platform training. Find out how contributing simple markdown tutorials can result in fantastically FAIR training.

New Feature: Automatic RMarkdown
28 January 2022
Further building on the work in the automatic Jupyter Notebook, we’ve now re-written the Jupyter export to be faster, and more importantly added support for R and RMarkdown! Check out an example material. Here we take the content of the tutorial, written like normal GTN markdown, and we automatically convert it to Jupyter Notebooks and now RMarkdown documents! Check out the documentation on how to setup your tutorials to support this.

New GTN Feature Tag-based Topics enables new SARS-CoV-2 topic
23 January 2023
The GTN has long struggled with the rigid hierarchy of a single level of classification: topics. All materials had to be organised into a single topic, even when their contents sometimes spanned multiple topics! We added subtopics a while back to help further organise these tutorials, but that still did not solve the issue of needing to collect multiple learn materials in a single location for people to access.

New Feature: Prometheus Metrics endpoint
24 January 2023
Prometheus is an increasingly popular monitoring solution whereby sites expose a /metrics endpoint, and then a central Prometheus server can scrape it for data which is then plotted or visualised or alerted upon. We at the GTN are now testing out a prometheus endpoint for our completely static site which we’ll use to expose some maybe useful statistics about the GTN that others can then scrape and display in ways that are useful for them.

GTN Celebrates Black History Month
2 February 2023
February marks Black History Month in the United States. This year we want to celebrate that by introducing a new theme (Click to Activate), and taking a look at one of the most famous Black figures in Science: Henrietta Lacks whose contribution to science went unremarked and unappreciated for decades while white scientists took credit for advancements built on her cell line.

What are the most used tools in the GTN?
8 February 2023
Did you ever wonder which was the number one most used tool in the GTN? There is a new page of Top Tools where we list all of the top tools in the GTN, and which tutorials use them. We guessed “upload1”, but it seems like not every author annotates that step explicitly in their tutorial, and of course it doesn’t appear in the workflows which we also scan to create this tool listing.

New Feature: Trainer Directory! (Add yourself today!)
21 March 2023
Numerous other training groups keep “Trainer Directories” as a way to help students meet trainers in their area, especially for hosting workshops for their local community. There have been a handful of attempts at this in the past, both in galaxyproject/galaxy-maps and in a Google Map on the GTN homepage.

New Feature: Click-to-run Workflows
19 April 2023
The GTN has implemented “click to run” workflows! One click, will get you into Galaxy with a workflow imported and ready to run.
If you are on either usegalaxy.org or usegalaxy.eu, we have implemented easy buttons which you can click that will direct you straight to your Galaxy server.

GTN in Discourse
30 January 2024
Howdy teachers and support staff! We are exploring displaying GTN content directly in the Galaxy Help site, furthering the integration between the GTN and the Galaxy community infrastructure.
When you paste a GTN Frequently Asked Questions URL into the Galaxy Discourse, it will now be rendered directly in Discourse!

GTN in Discourse
30 January 2024
Howdy teachers and support staff! We are exploring displaying GTN content directly in the Galaxy Help site, furthering the integration between the GTN and the Galaxy community infrastructure.
When you paste a GTN Frequently Asked Questions URL into the Galaxy Discourse, it will now be rendered directly in Discourse!

Cool URLs Don't Change, GTN URLs don't either.
18 March 2024
At the Galaxy Training Network we are really committed to ensuring our training materials are Findable, Accessible, Interoperable, and Reusable.
This means that we want to make sure that the URLs to our training materials are persistent and don’t change.
The GTN wants you to be able to rely on our URLs once you’ve added them to a poster or training material, without having to worry about them breaking in the future.

Cool URLs Don't Change, GTN URLs don't either.
18 March 2024
At the Galaxy Training Network we are really committed to ensuring our training materials are Findable, Accessible, Interoperable, and Reusable.
This means that we want to make sure that the URLs to our training materials are persistent and don’t change.
The GTN wants you to be able to rely on our URLs once you’ve added them to a poster or training material, without having to worry about them breaking in the future.

Learn to use MINERVA Platform's COVID-19 Disease Map with Galaxy
28 March 2024
As part of the work of BeYond COVID, we have developed a new integration between the MINERVA Platform’s COVID-19 Disease Map and Galaxy! Datasets created within Galaxy can now be seamlessly visualized in the MINERVA Platform, allowing you to explore your data in the context of the COVID-19 Disease Map.

Learn to use MINERVA Platform's COVID-19 Disease Map with Galaxy
28 March 2024
As part of the work of BeYond COVID, we have developed a new integration between the MINERVA Platform’s COVID-19 Disease Map and Galaxy! Datasets created within Galaxy can now be seamlessly visualized in the MINERVA Platform, allowing you to explore your data in the context of the COVID-19 Disease Map.
GTN Video Library 2.0: 107 hours of learning across 154 videos
14 June 2024
Many GTN tutorials already have recordings. These recordings were made by members of the community for a variety of (online) training events.
Up until now, this video library were part of the Gallantries Project.
We have now integrated this video library directly into the GTN, and made it even easier to add video recordings to GTN tutorials or slide decks! Just use a Google Form to submit your video recordings!

GTN Video Library 2.0: 107 hours of learning across 154 videos
14 June 2024
Many GTN tutorials already have recordings. These recordings were made by members of the community for a variety of (online) training events.
Up until now, this video library were part of the Gallantries Project.
We have now integrated this video library directly into the GTN, and made it even easier to add video recordings to GTN tutorials or slide decks! Just use a Google Form to submit your video recordings!
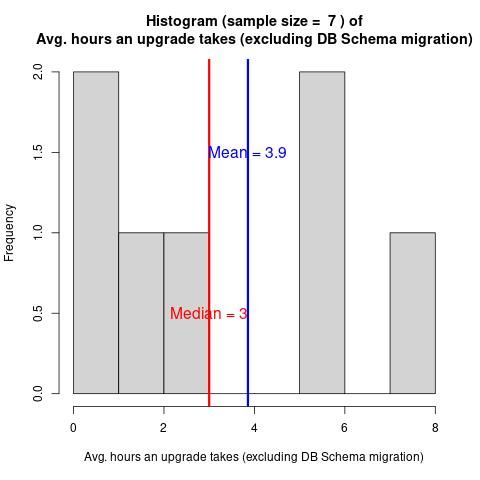
Galaxy Administrator Time Burden and Technology Usage
22 July 2024
Have you wondered how difficult Galaxy is to run? How much time people must spend to run Galaxy?
In February 2024, we collected 9 responses from the Galaxy Small Scale Admin group.
The questions cover various time burdens and technological choices.
The report provides answers to prospective future admins’ most common questions.
Credit where it's due: GTN Reviewers in the spotlight
2 December 2024
We would like to recognise and thank all of the reviewers who have contributed to the GTN tutorials; your efforts are greatly appreciated, and we are grateful for your contributions to the GTN community. Today, we are highlighting your efforts on every single learning material across the GTN.

Credit where it's due: GTN Reviewers in the spotlight
2 December 2024
We would like to recognise and thank all of the reviewers who have contributed to the GTN tutorials; your efforts are greatly appreciated, and we are grateful for your contributions to the GTN community. Today, we are highlighting your efforts on every single learning material across the GTN.
External Links

Favourite Topics
Favourite Formats